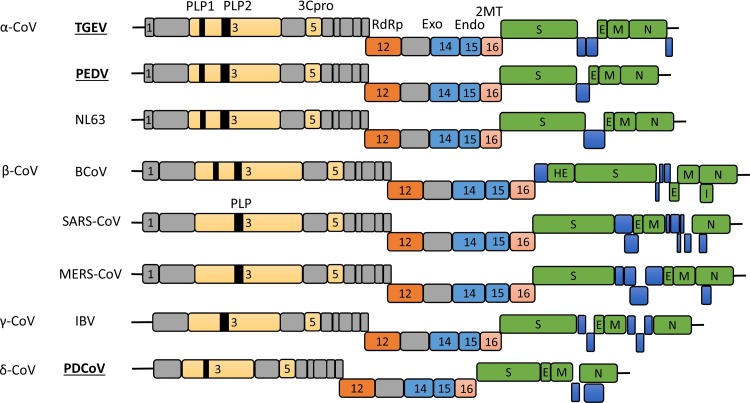
Fig. 1.
Comparative genome organization of the representative coronaviruses from each genus in the family Coronaviridae. Swine enteric coronaviruses (TGEV, PEDV, and PDCoV) are indicated in bold underlined. Structural proteins are presented in green. Putative accessory proteins are shown in blue. Nonstructural proteins in ORF1a/b are presented in grey or other colors. Papain-like proteinases (PLP1 and PLP2), coronavirus 3C-like proteinase (3Cpro), RNA-dependent RNA polymerase (RdRp), two viral nucleases (nsp14 as exonuclease and nsp15 as endonuclease), and 2′-O-methytransferase (2MT; nsp16) are indicated by respective nsp numbers. The genome for Alaphacoronavirus and Betacoronavirus contains the nsp1 gene in different lengths, whereas the genome for Gammacoronavirus and Deltacoronavirus lacks the nsp1 gene. TGEV, transmissible gastroenteritis coronavirus; PEDV, porcine epidemic diarrhea virus; NL63, human coronavirus NL63; BCoV, bovine coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, middle east respiratory syndrome coronavirus; IBV, avian infectious bronchitis virus; PDCoV, porcine deltacoronavirus; HE, hemagglutinin-esterase; S, spike; E, envelope; M, membrane; N, nucleocapsid; I, internal open reading frame.