Fig. 1.
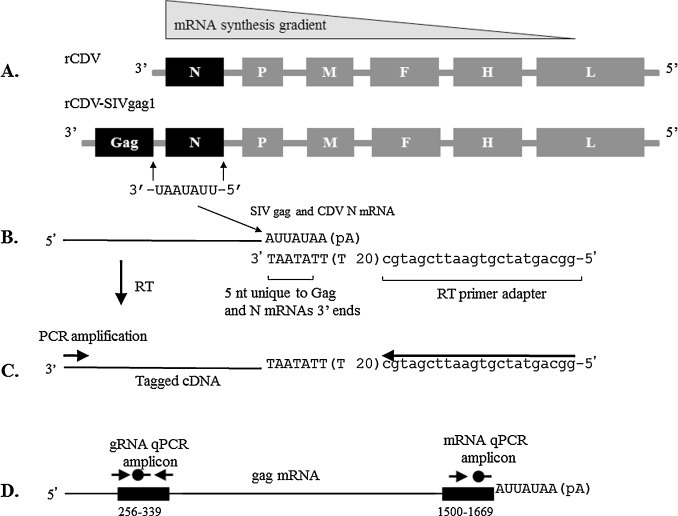
The relative frequency of mRNA transcription, genome organization for each vector, the scheme for the mRNA specific RT-qPCR, and the binding sites for qPCR primers and probe for the SIV gag gRNA and mRNA assays are depicted. (A). Transcriptional attenuation resulting in a gradient of mRNA synthesis such that the gag message is more abundant than the N message in the rCDV-SIVgag1 vector; the rCDV vector is a Vero cell adapted, Onderstepoort live-attenuated CDV vaccine (Galaxy D, Schering-Plough Animal Health Corporation, Omaha, NE, USA) obtained through a commercial source; and the rCDV-SIVgag1 vector with the Gag gene in the first position of the genome and a unique UAUAAU sequence common to CDV N and SIV gag genomes. (B). Diagram of the RT step used in the singleplex or duplex assay to synthesize cDNA from mRNA. The CDV-specific mRNA RT-primer that binds to a unique UAUUA sequence common only to CDV N and SIV gag mRNA transcripts, has a 22-base adapter sequence at the 5′ end followed by a 20-base oligo-dT sequence, and a 3′-terminal 5-base anchor (ATAAT). (C) Tagged cDNA generated with this primer is then PCR amplified with CDV N and SIV-gag forward primers and labeled probes and the CDV sequence tag-reverse primer that corresponds to the adapter sequence, as described in Section 2. (E). The black box from 256–339 Nt indicates the binding site for the qPCR primers ( ) and probe (
) and probe ( ) for the Gag specific gRNA RT-qPCR assay and the 1500–1669 Nt indicates the amplicon and the binding site for qPCR forward primer (
) for the Gag specific gRNA RT-qPCR assay and the 1500–1669 Nt indicates the amplicon and the binding site for qPCR forward primer ( ) and probe (
) and probe ( ) used in the Gag specific mRNA RT-qPCR assay.
) used in the Gag specific mRNA RT-qPCR assay.
