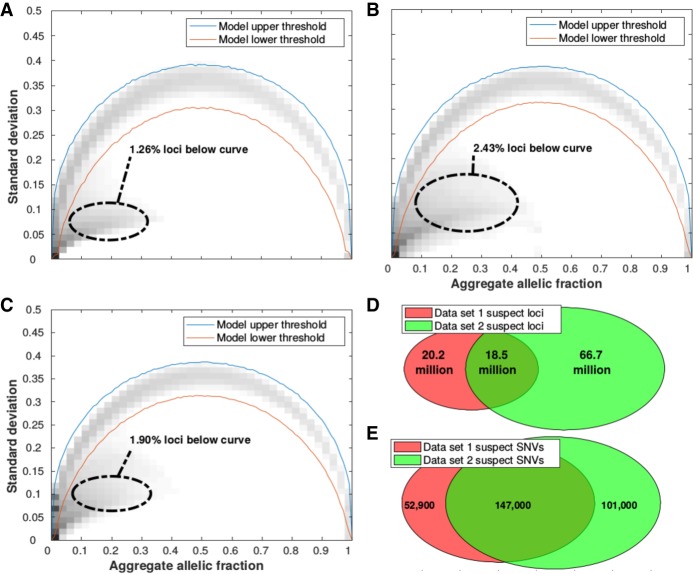
Figure 2.
Identification of suspect autosomal loci/allele combinations with persistent low allelic fractions across patients. Observed and expected alternative allele fractions were estimated from five different whole-genome sequenced cohorts: (A) Data set 1, (B) Data set 2, (C) Data set 3. For all loci in autosomal chromosomes, the standard deviation and aggregate allelic fraction values from the IncDB were plotted against each other in a density plot. The darker regions have the highest concentration of loci. The red lines indicate the upper and lower boundaries of the 99.9% confidence interval for the expected allelic standard deviation (shown in Supplemental Fig. S1A,B, respectively). Suspect loci for each cohort were defined as the loci with standard deviation below their simulated model lower threshold. A total of 2.8 billion autosomal loci were assessed. (D) Venn diagram showing the overlap of all suspect loci between data sets 1 and 2. (E) Venn diagram of overlap at suspect loci where SNVs have been called. A total of 3.44 million autosomal SNVs were not annotated as suspect in either data set 1 or 2.