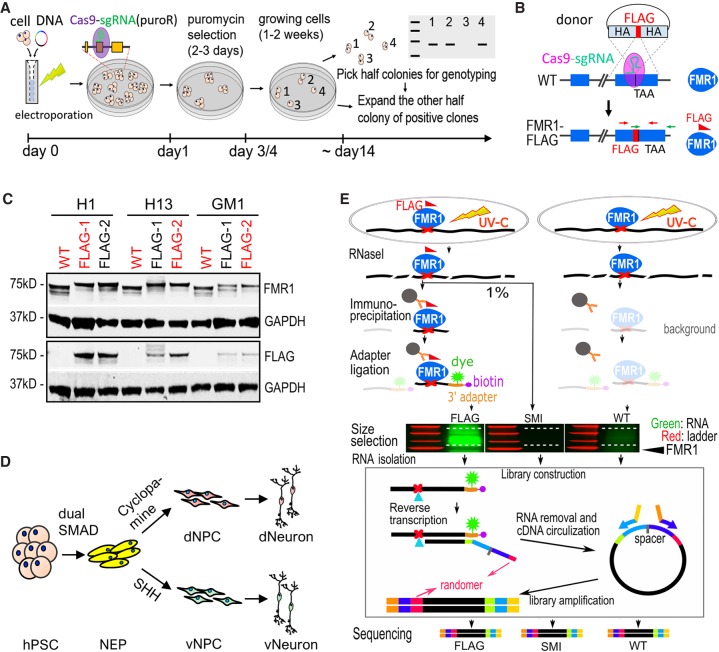
Figure 1.
Generation of FMR1-FLAG hPSCs and neural cells for CLIP. (A) Schematic diagram of one-step seamless genome editing using CRISPR-Cas9. A Cas9-sgRNA plasmid also confers puromycin resistance (puroR) which allows for a temporary selection to obtain seamless genome editing in ∼2 wk. (B) Diagram of generation of FMR1-FLAG hPSCs using CRISPR-Cas9 and a donor plasmid for FLAG knock-in. Exons of FMR1 are shown as blue boxes. TAA indicates the position of a stop codon. HA represents homology arm, and crossed dash lines depict homology-directed recombination. (C) Western blot detecting FMR1 or FLAG in FMR1-FLAG and WT hPSCs, GAPDH as a loading control. hPSC lines in red were used for experiments. (D) Diagram of neural differentiation of hPSCs. Neuroepithelial cells (NEP) were differentiated from hPSCs by dual SMAD inhibition followed by patterning to forebrain dorsal NPCs (dNPC) and ventral MGE-like NPCs (vNPC) by modulating the SHH pathway. Patterned NPCs were further differentiated to neurons. (E) Flow chart of CLIP-seq. Both FMR1-FLAG and control (WT) cells were subjected to UV crosslinking. RNAs were partially digested by RNase I and FMR1-bound RNAs were immunoprecipitated using a FLAG antibody. Immunoprecipitated RNAs (and background levels of RNAs) were ligated to a fluorescent 3′ adapter for visualization (green) and reverse transcription. Randomers were incorporated in the RT primer to allow for removal of duplicated Illumina reads from individual cDNAs. A size-matched input (SMI) control, RNAs from FMR1-FLAG cells that were not immunoprecipitated but size-selected, was also used as a control.