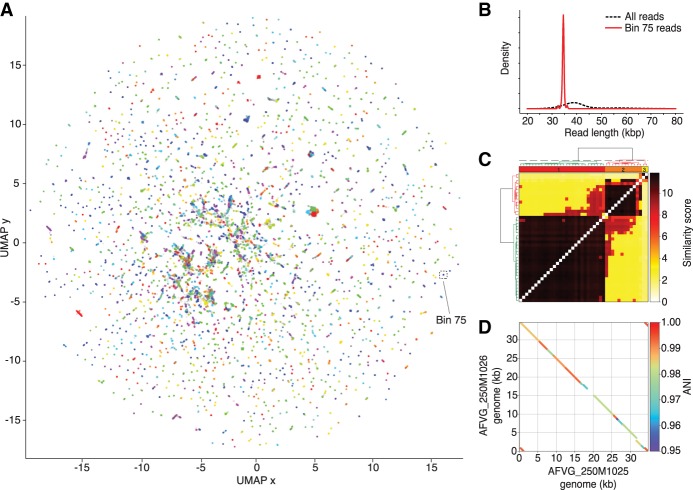
Figure 2.
Phage discovery steps for 250-m sample. (A) Nanopore reads that were found to contain DTRs were first represented by 5-mer count vectors and dimensionally reduced into a 2D embedding using UMAP (McInnes et al. 2018). Reads in the 2D embedding are colored based on their assignment to the 2386 5-mer bins called by hdbscan (McInnes et al. 2017). 5-mer bin colors are redundant owing to the large number of bins. Reads not assigned to a bin are colored gray. The 5-mer bin 75 is highlighted and contains 42 nanopore reads. (B) Read lengths from bin 75 were compared with read lengths from all bins, revealing an enrichment of ∼35-kb reads and a depletion of reads at other lengths. (C) Reads within bin 75 were aligned to each other to generate pairwise alignment scores, which were hierarchically clustered to reveal two main alignment clusters. A polished assembly-free virus genome (AFVG) was generated from each cluster: AFVG_250M1025 and AFVG_250M1026. (D) Comparison between the AFVG_250M1025 and AFVG_250M1026 sequences shows microdiversity between the two polished phage genomes. The genomes share large regions of varying degrees of sequence identity >95% with interspersed divergent sequence.