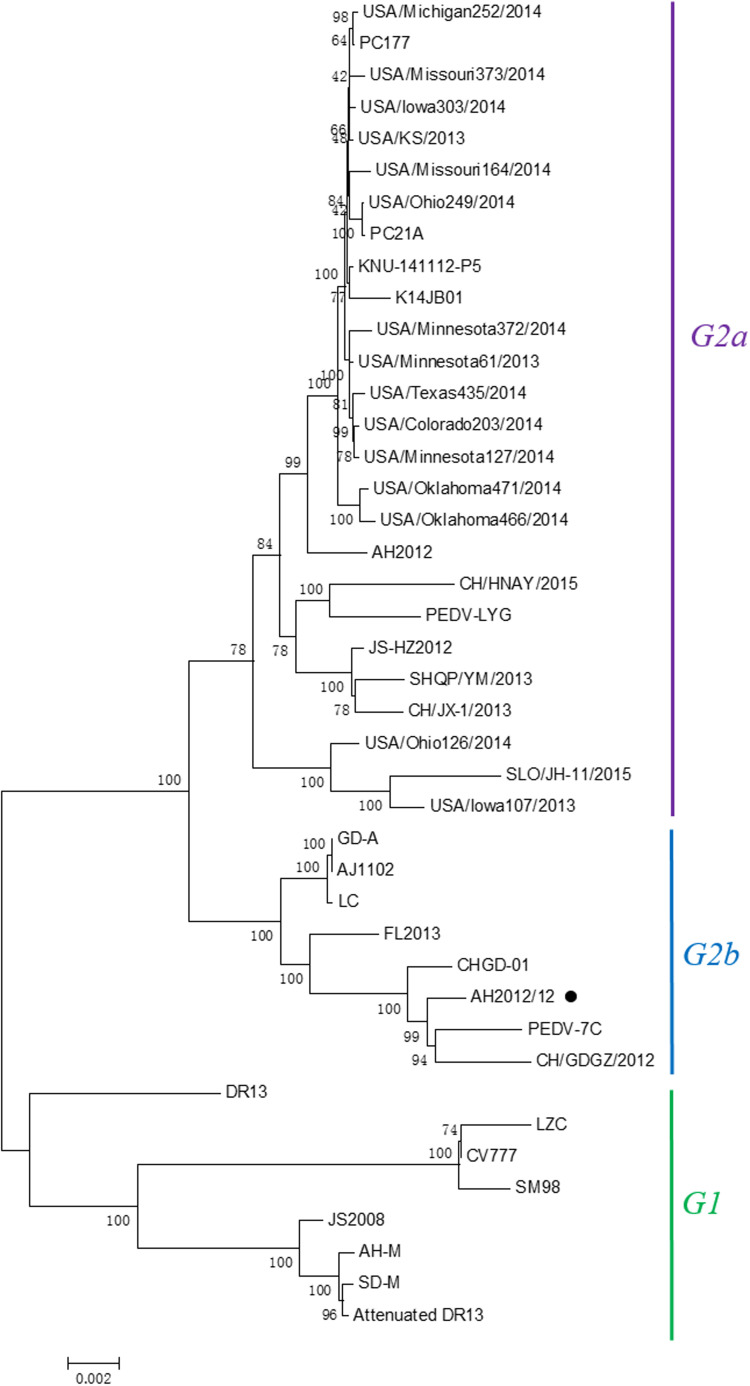
Fig. 3.
Phylogenetic analysis based on the nucleotide sequence of the full-length genome of AH2012/12. The evolutionary history was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) were shown next to the branches. The evolutionary distances were computed using the Jukes-Cantor method and were presented as the number of base substitutions per site. Evolutionary analyses were conducted using MEGA5 software.