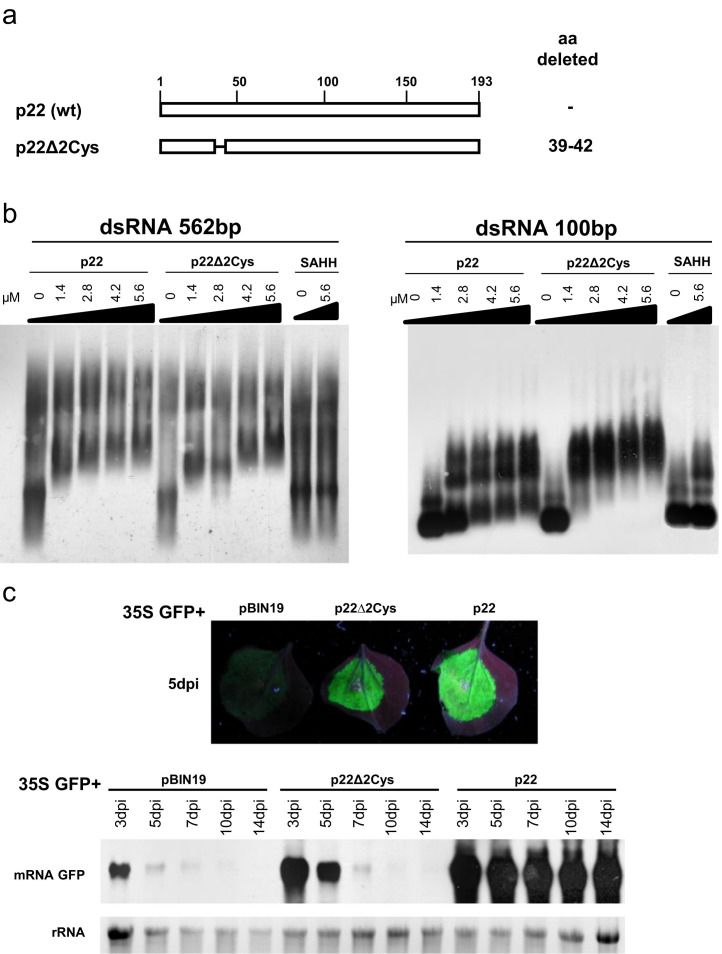
Fig. 4.
Analysis of RNA-binding properties and silencing suppression activity of Tomato chlorosis virus -p22 deletion mutant. (a) Schematic representation of the wild-type p22 protein and the deletion mutant. The deleted amino acid (aa) residues are indicated. (b) RNA-binding properties of the p22Δ2Cys mutant detected by an EMSA assay. The increased concentration of protein is indicated above each lane. Wild-type p22 was used as a positive control, and S-adenosylhomocysteine hydrolase (SAHH) was used as a negative control. The EMSA gels were transferred to a nylon membrane, and dsRNAs were detected using an anti-digoxigenin antibody and a chemiluminescent substrate. (c) Silencing suppression assays with a 35S construct expressing the p22Δ2Cys mutant. Upper panels show photographs taken under UV light of Nicotiana benthamiana leaves at 5 days postinfiltration (dpi) with Agrobacterium tumefaciens harboring 35S GFP in combination with the empty vector pBIN19 or with a 35S construct expressing p22Δ2Cys or with p22. The lower panels show a northern blot analysis of GFP mRNA extracted from the zones infiltrated at 3, 5, 7, 10 and 14 dpi, hybridized with a probe specific to GFP mRNA. Ethidium bromide staining of rRNA was used as a loading control.