Fig. 1.
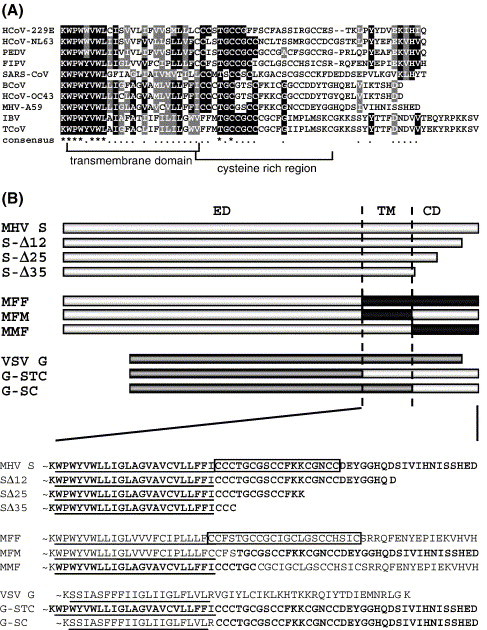
(A) A CLUSTALW alignment of the carboxy-terminal spike protein sequences from nine coronaviruses including feline infectious peritonitis virus (FIPV, strain 79-1146; GenBank accession no. VGIH79), porcine transmissible gastroenteritis virus (TGEV, strain Purdue; GenBank accession no. P07946), porcine epidemic diarrhea virus (PEDV; GenBank accession no. NP_598310), HCoV-229E (human coronavirus, strain 229E; GenBank accession no. VGIHHC), bovine coronavirus (BcoV, strain F15; GenBank accession no. P25190), mouse hepatitis virus (MHV, strain A59; GenBank accession no. P11224), HCoV-OC43 (human coronavirus, strain OC43; GenBank accession no. CAA83661), SARS-CoV (strain TOR2; GenBank accession no. P59594) and infectious bronchitis virus (IBV, strain Beaudette; GenBank accession no. P11223). The transmembrane domain and cysteine-rich region are indicated. (B) Diagrams of wild-type MHV S protein; cytoplasmic domain truncation mutants SΔ12, SΔ25, and SΔ35; MHV-FIPV chimeras MFF, MFM, and MMF; and VSV G wild-type and VSV-MHV chimeric proteins G-STC and G-SC. Light grey boxes represent the MHV S amino acids sequences, black boxes indicate FIPV S sequences, whereas the dark grey bars represent sequences of the VSV G. ED, ectodomain; TM, transmembrane domain; CD, cytoplasmic domain. Below, TM and CD amino acid sequences of all constructs shown above. The cysteine-rich region in the MHV S and MFF protein has been boxed. The MHV S-derived sequences are indicated in bold. TM regions are underlined.
