Fig. 2.
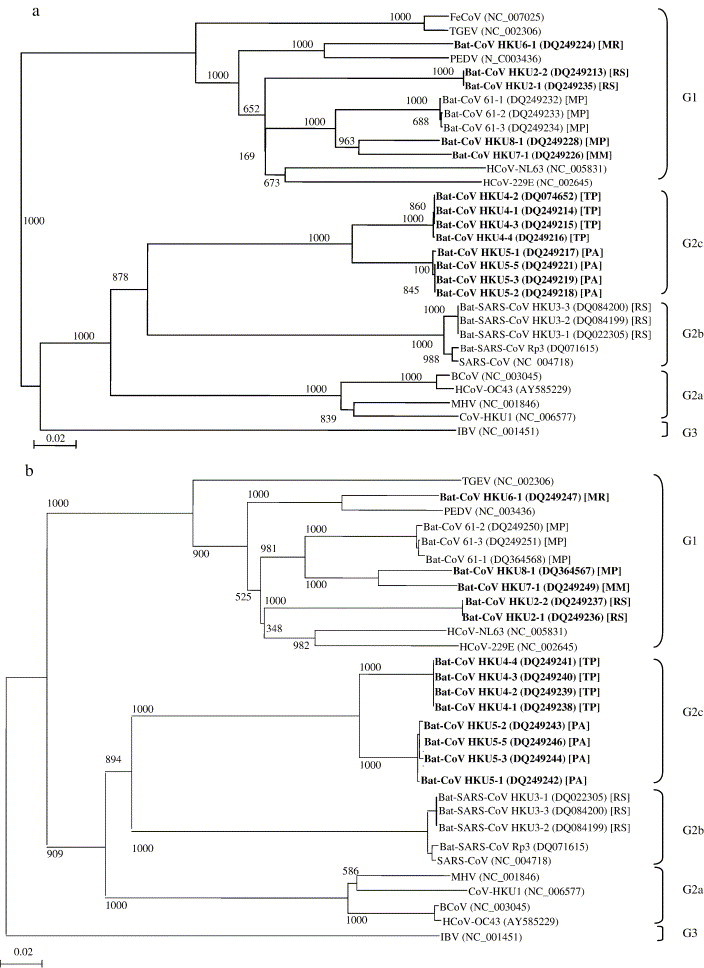
Phylogenetic trees of Pol (2a) and helicase (2b) showing the relationship of the novel bat coronaviruses to known coronaviruses. The trees were inferred from amino acid sequence data (949 amino acid positions for RNA-dependent RNA polymerase and 609 amino acid positions for helicase) by the neighbor-joining method. Numbers at nodes indicated levels of bootstrap support calculated from 1000 trees. The scale bar indicates the estimated number of substitutions per 50 amino acids. Only three of the 21 strains of bat-SARS-CoV are shown. GenBank accession numbers are in brackets and names of bats from which the coronaviruses were recovered are in square brackets. The 13 strains of the six novel coronaviruses in bats are in bold. HCoV-229E, human coronavirus 229E; PEDV, porcine epidemic diarrhea virus; TGEV, porcine transmissible gastroenteritis virus; FeCoV, feline coronavirus; HCoV-NL63, human coronavirus NL63; HCoV-OC43, human coronavirus OC43; MHV, murine hepatitis virus; BCoV, bovine coronavirus; CoV-HKU1, coronavirus HKU1; SARS-CoV, SARS coronavirus; bat-SARS-CoV, SARS coronavirus-like virus found in bats; IBV, infectious bronchitis virus; MR, Myotis ricketti; RS, Rhinolophus sinicus; MP, Miniopterus pusillus; MM, Miniopterus magnater; TP, Tylonycteris pachypus; PA, Pipistrellus abramus.
