Supplementary Fig. 1.
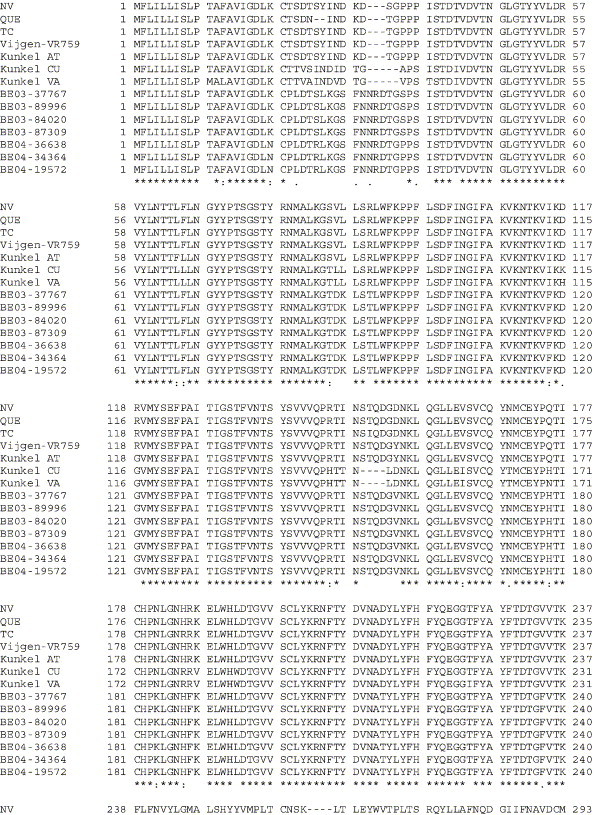
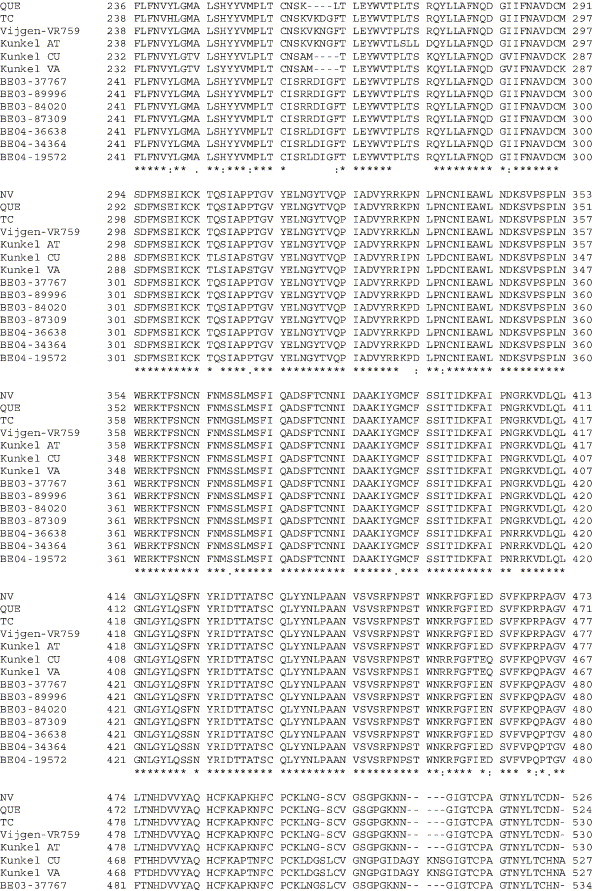
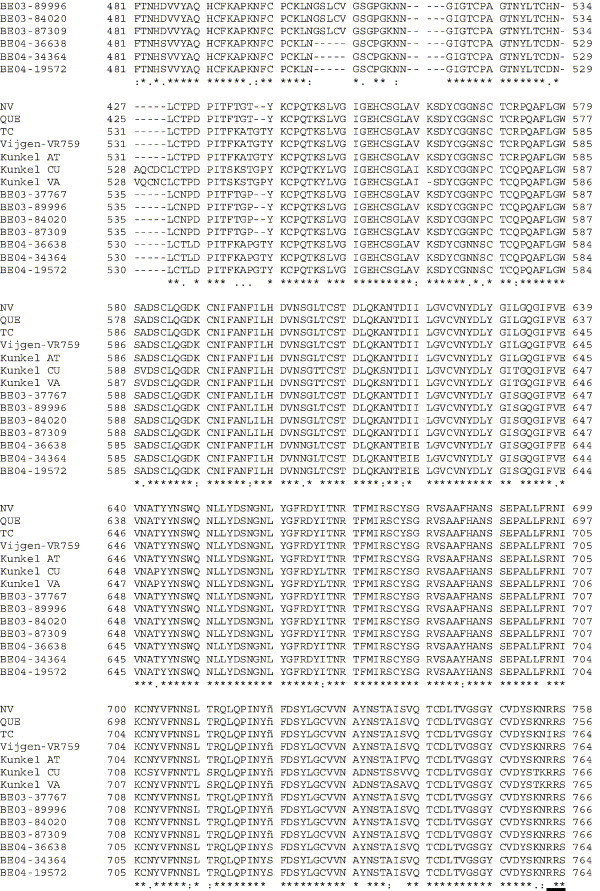
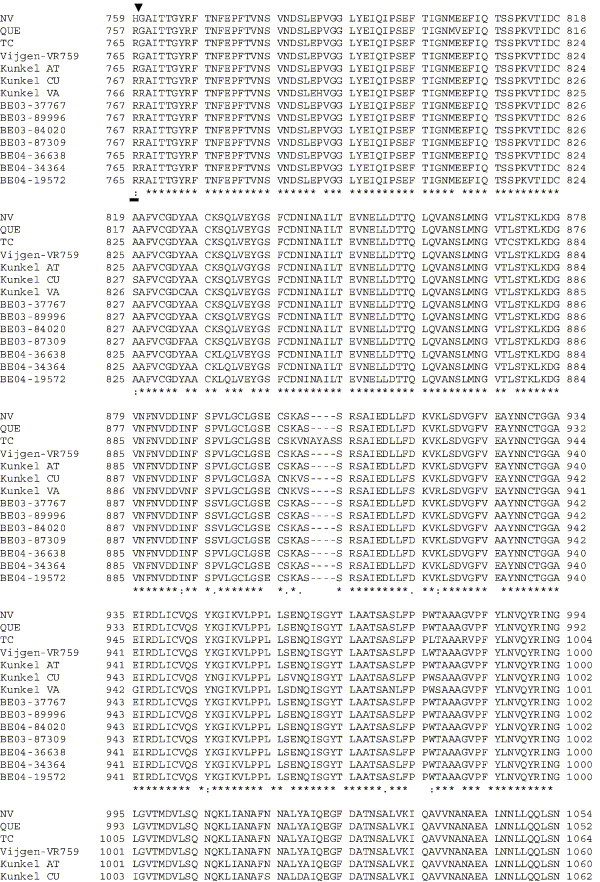
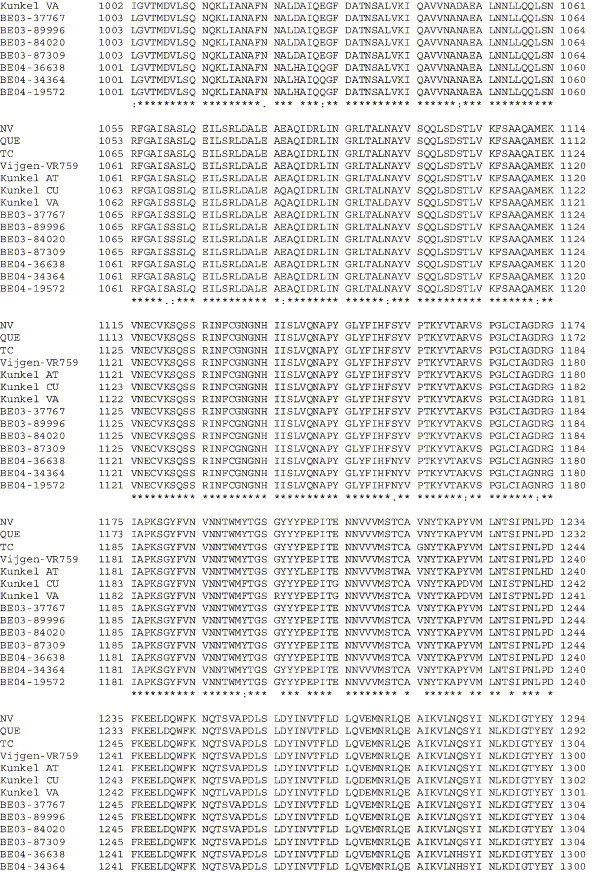
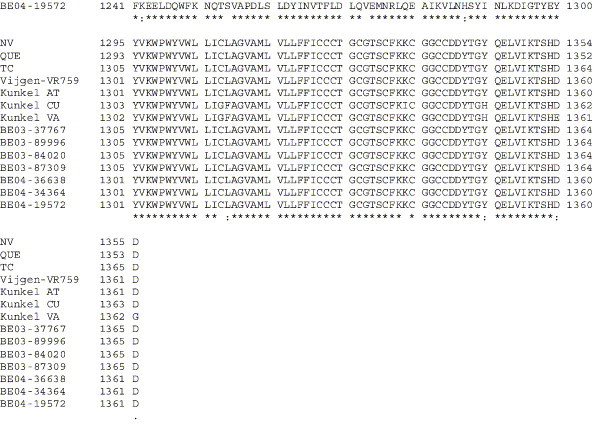
Amino acid alignment of S protein sequences for HCoV-OC43NV, HCoV-OC43QUE, HCoV-OC43TC and the published sequence of several HCoV-OC43 isolates. Asterisks indicate identical residues, colons represent conserved changes and blank spaces denote non-conserved substitutions. The putative furin recognition site is underlined and the cleavage site is marked with an arrowhead. For HCoV-OC43NV and HCoV-OC43TC, total RNA was reverse transcribed to cDNA and the S gene was amplified by PCR. PCR products were directly sequenced as described in Materials and methods. The sequence for HCoV-OC43QUE corresponds to GenPept accession number AAT84362, passed 5–6 times in HRT-18 cells (St-Jean et al., 2004); the sequence for Vijgen VR-759 was translated from GenBank accession number AY391777, passed an unknown number of times in HRT-18 cells (Vijgen et al., 2005b); the sequence for Kunkel AT corresponds to GenPept accession number AAB27260, passed 3 times in HRT-18 cells (Kunkel and Herrler, 1993b); the sequences for Kunkel CU and Kunkel VA correspond to GenPept accession numbers CAA83660 and CAA83661, passed 3 times in MDCK cells and 12 times in Vero E6 cells, respectively (Kunkel and Herrler, 1996); and sequences for HCoV-OC43 clinical isolates BE03-37767, BE03-89996, BE03-84020, BE03-87309, BE04-36638, BE04-34364 and BE04-19572 correspond to GenPept accession numbers AAX84794, AAX84791, AAX84793, AAX85674, AAX84795, AAX84792 and AAX85678, respectively, submitted by Vijgen et al. (2005a).
