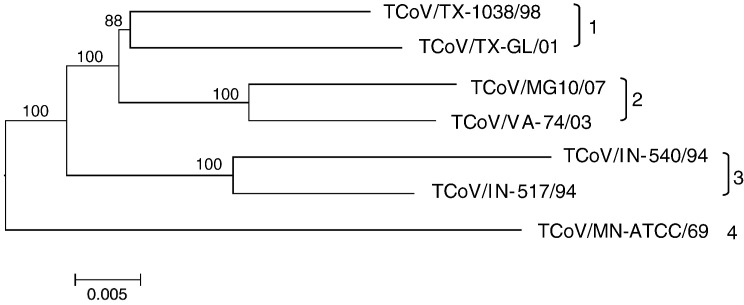
Fig. 2.
Neighbor-Joining method used to infer evolutionary history using full genomic sequence data for available TCoV isolates. The bootstrap consensus sub-tree was constructed from 1000 replicates (percentage of replicate trees in which associated strains clustered together are presented at nodes). The p-distance scale is presented at the bottom of the figure. Different genetic groups are indicated at the right of the figure.