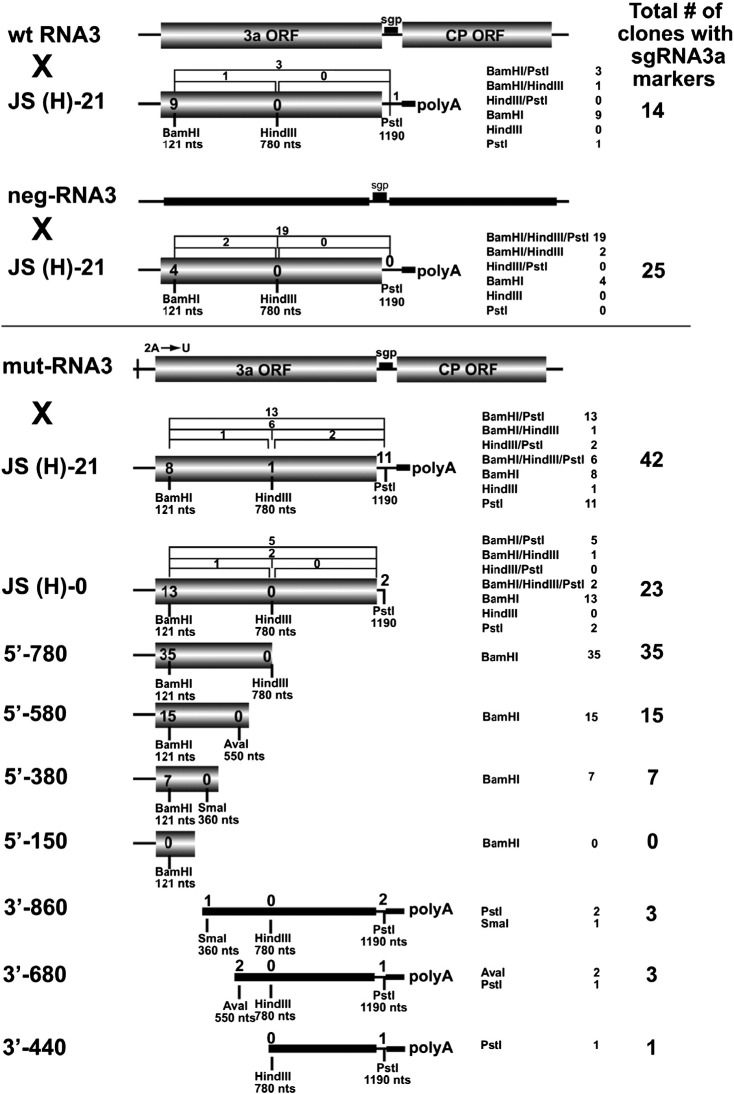
Fig. 5.
Recombination of BMV RNA3 templates with sgRNA3a constructs in barley protoplasts. wt RNA3 × JS(H)-21 indicates an experiment with wt genomic RNA3 (wt RNA3) and full-length sgRNA3a (JS(H)-21). neg-RNA3 × JS(H)-21 indicates an experiment with negative-strand RNA3 (neg-RNA3) and full-length sgRNA3a (JS(H)-21). mut-RNA3 × JS(H)-21 indicates an experiment with an RNA3 derivative bearing a point mutation (A → U) at position 2 near the 5′ end. The remaining sgRNA3a-derivatives that were tested for recombination with mut-RNA3 are shown below, including JS(H)-0: an sgRNA3a derivative without a polyA tail; 5′-780, 5′-580, 5′-380, 5′-150: the 5′-nested deletion sgRNA3a fragments; and 3′-860, 3′-680, 3′-440: the 3′-nested deletion sgRNA3a fragments. The positions of marker restriction sites are indicated below each construct. The column on the right shows the general recombination frequency (per-cent) based on an analysis of one-hundred insert-bearing cDNA clones. The frequency of recombinants bearing single restriction markers is shown inside the shaded rectangles, whereas that of those carrying double or triple markers is represented above the constructs as brackets. The 3′nested RNAs do not carry the 3a ORF initiation codon and they are therefore represented by thick black lines. The other elements are as in Fig. 1. The numbers summarize the results from two independent transfection experiments with each sgRNA3a derivative.