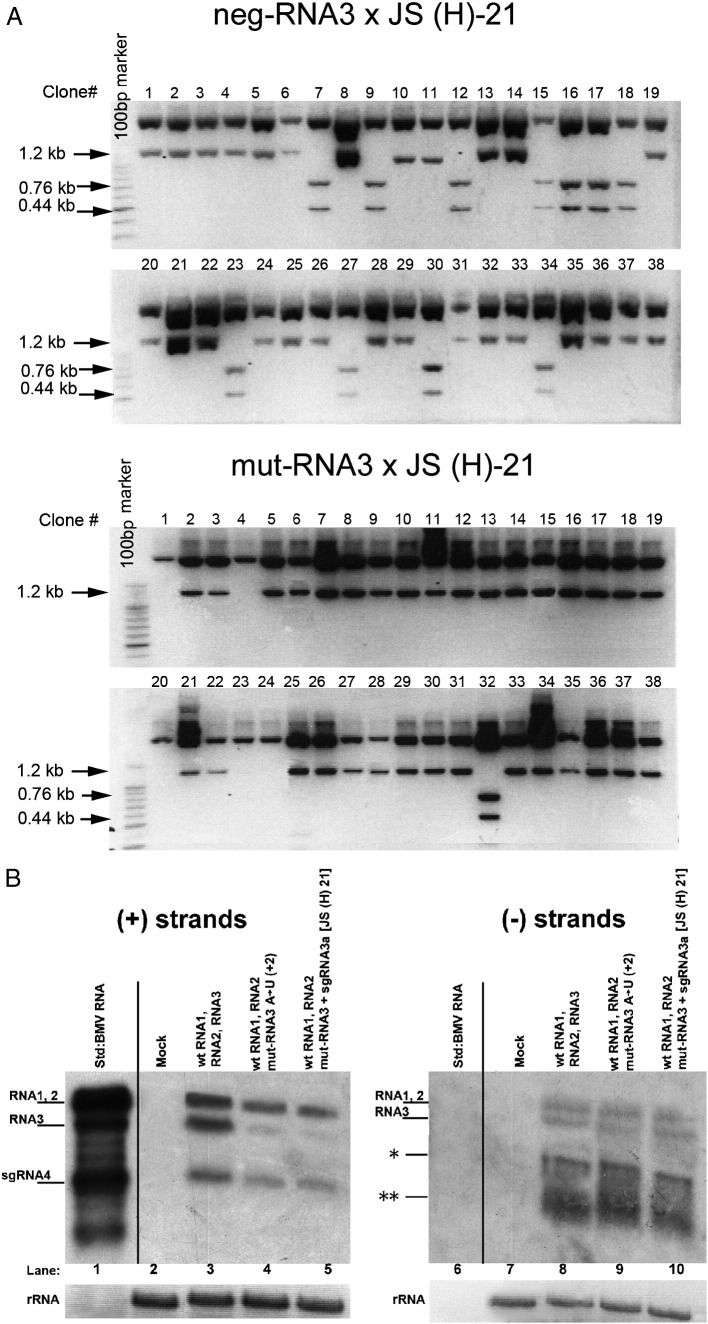
Fig. 6.
Recombination of BMV RNA3 with sgRNA3a in barley protoplasts. A. Restriction enzyme digestion of recombinant RNA3 cDNA clones obtained from barley protoplasts that were transfected with a mixture of wt BMV RNAs 1 and 2 and (top) Neg-RNA3 plus JS(H)-21 sgRNA3a or (bottom) mut-RNA3 and JS(H)-21 sgRNA3a. The RNA3 region representing the sgRNA3a sequence was amplified from total RNA extracts by RT-PCR, and the cDNA products were cloned into the pGEM-T Easy system. The insert sequences were released by HindIII/EcoRI digestion and separated by electrophoresis in a 1.5% agarose gel. An intact 1.2-kb fragment reflected the lack of the HindIII marker site at nt position 780, whereas the double 0.44 kb and 0.76 kb bands indicate the presence of the HindIII marker. B. Northern blot analysis showing the accumulation in protoplasts of either (+) or (−) strands (left and right panels, respectively) of BMV RNAs. Total protoplast RNA was separated in a 1.2% denaturing agarose gel, blotted, and probed with a 3′-specific RNA probe detecting either (+) or (−) strands (see Materials and methods). Lanes 1 and 6, virion BMV RNA used as size standards; lanes 2 and 7, negative controls from mock inoculated protoplasts; lanes 3 and 8, protoplasts transfected with equimolar amounts of BMV RNAs 1 and 2 and wt RNA3; lanes 4 and 9, protoplasts transfected with BMV RNAs 1 and 2 and (A → U) RNA3; lanes 5 and 10, protoplasts transfected with BMV RNAs 1 and 2, (A → U) RNA3, and sgRNA3a. Ribosomal RNA (rRNA) bands (after staining with ethidium bromide) are shown below. The position corresponding to the (−) RNA4 band is marked with a single asterisk on the right panel, whereas the migration position of degradation products (likely because minus strands are not encapsidated and thus less protected) is marked with two asterisks.