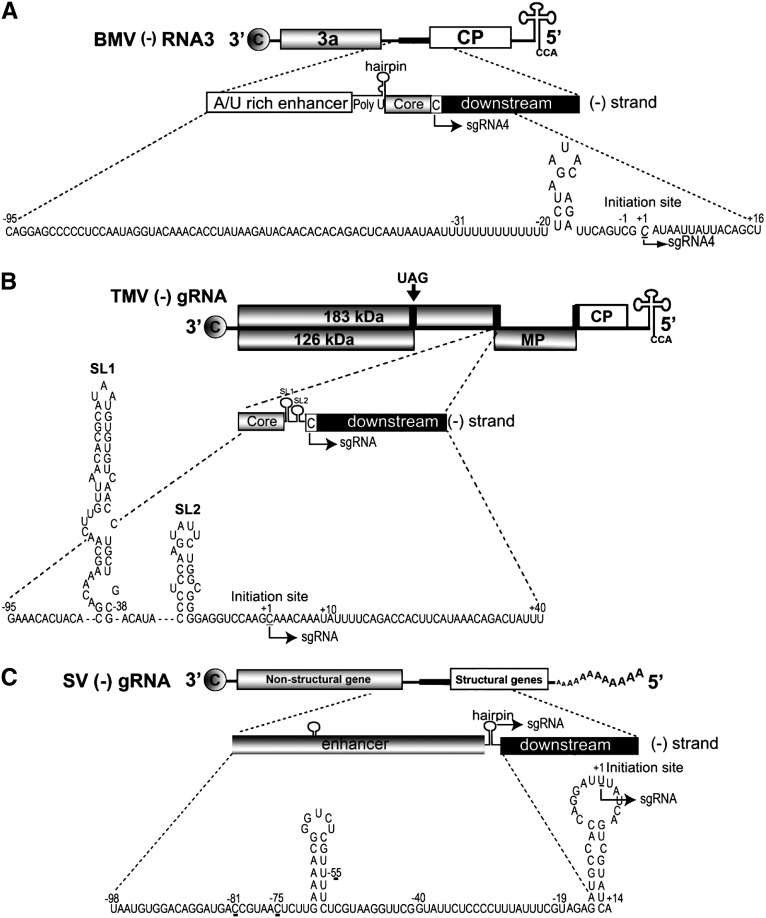
Fig. 2.
Schematic organization of subgenomic (SGP) promoters in RNA viruses. (A) The SGP domain is shown as a solid, black box located between the 3a and CP ORFs on BMV (−) RNA3. The arrow indicates the initiation site and the direction of subgenomic RNA4 synthesis. The bottom expansion shows the nucleotide sequence of the SGP promoter (− 95 to + 16). The SGP subdomains are indicated as follows: the enhancer including the poly U tract (nt − 95 to − 20), the core region (nt − 19 to − 1) including the hairpin, the initiation + 1 cytidilate, and the downstream portion. (B) TMV SGP promoters are indicated by the black rectangles: the first promoter is located between 183-kDa RDRP protein ORF and MP ORFs, and the second one between MP and CP ORFs. The arrow above the G RNA indicates the position of the amber read-through codon, the putative initiation site for I1 SG RNA expressing 54-kDa protein of unknown function. The arrow below (−) G RNA indicates the initiation site for one of the SG RNAs. The bottom expansion shows the nucleotide sequence of the SGP promoter (− 95 to + 40). The promoter core (− 35 to + 10),the two hairpin structures, SL1 and SL2, are indicated,as well as the initiation + 1 cytidilate, and the downstream portion (Grdzelishvili et al., 2000). (C) The SGP promoter of SV (−) strand RNA is shown in the top line as a solid, black box. The arrow on (−) G RNA diagram indicates the initiation site and the direction of SG RNA synthesis. The bottom expansion shows the nucleotide sequence of the SGP promoter (− 98 to + 14). The sequence between − 19 to + 5 represents the minimal sequence with promoter activity. The sequence from − 40 to + 14 was shown to enhance the promoter activity. Mutations at the marked nucleotide positions: − 81, − 75, − 55, down regulate the SGP promoter activity.