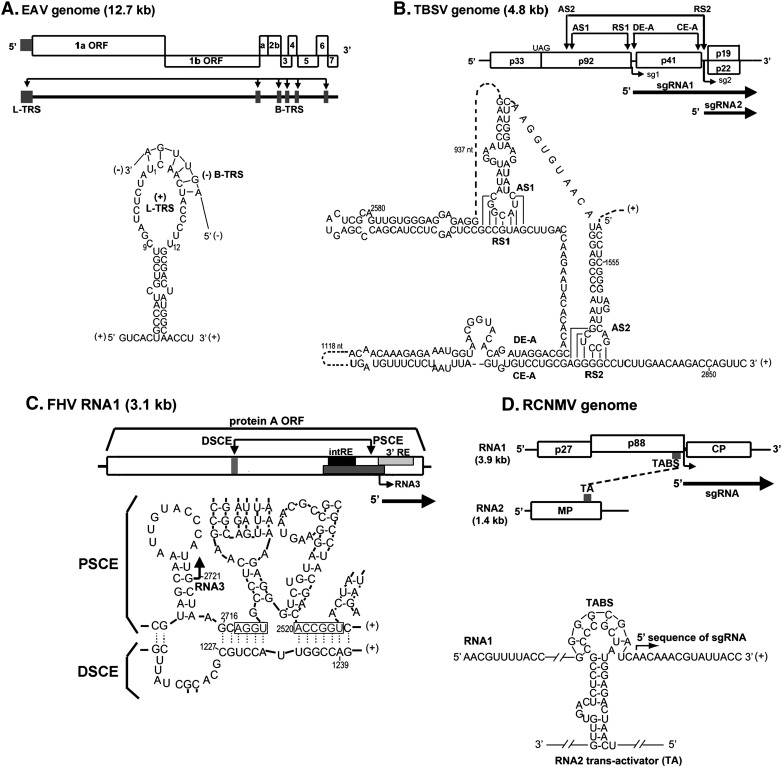
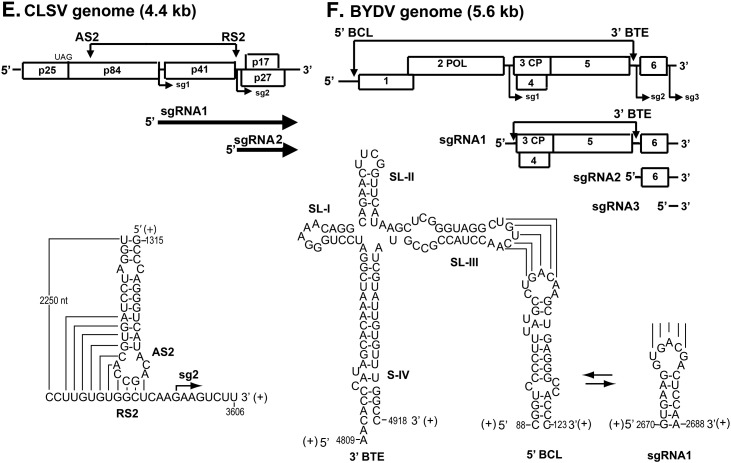
Fig. 3.
Long distance RNA–RNA interactions that regulate SG RNA synthesis in RNA viruses. (A) Schematic overview of the genome organization and expression of EAV. The gray boxes in the genomic RNA indicate the positions of the leader TRS and (major) body TRSs. Below is shown the proposed base pairing interaction between the sense leader TRS (L-TRS) and the antisense body TRS (B-TRS) in the 3′ end of the nascent minus strand (van Marle et al., 1999); only the loop and the top of the stem of the predicted hairpin structure are presented (B) Linear representation of the TBSV RNA genome showing its coding organization. The relative positions of interacting RNA elements involved in SG RNA transcription are shown above the genome and are indicated by arrows. Initiation sites for SG RNA transcription are labeled sg1 and sg2, and corresponding structures of the two SG RNAs are represented by bold arrows below the genome. Below, the long-distance RNA-RNA interactions that regulate SG RNA transcription in TBSV are shown in detail. Relevant sequences of the TBSV genome are presented with corresponding genomic coordinates. The AS1/RS1 base-pairing interaction is essential for the efficient transcription of SG RNA1 (Choi and White, 2002), while the AS2/RS2 and DE-A/CE-A base-pairing interactions promote SG RNA2 transcription (Choi et al., 2001, Lin and White, 2004); (C) Representation of genomic RNA1 of Flock house virus (FHV). Interaction between the distal subgenomic control element (DSCE) and the proximal subgenomic control element (PSCE) is required for RNA3 synthesis. The internal replication element (intRE) and 3′ replication element (3′ RE) are required for RNA1 replication. Below, potential base pairing of the DSCE to regions proximal to the subgenomic region start site. Gray boxes mark PSCE residues that potentially base pair to the DSCE. Putative helices 1 and 2 are bracketed. Arrow, RNA3 start site at nt 2721 (Lindenbach et al., 2002); (D) Schematic of the RCNMV genome showing the relative positions of the in trans interacting RNA elements: the loop portion of a stem-loop in RNA2 (termed trans-activator or TA) and a complementary sequence in RNA1 (termed TA binding site or TABS) located just upstream from the initiation site for SG mRNA transcription (Guenther et al., 2004); (E) Representation of CLSV genome and the proposed AS2 and RS2 interaction during regulation of SG RNA2 synthesis (Xu and White, 2008); (F) Overview of the BYDV genome and the in cisinteraction between genomic 5′-UTR stem-loop (BCL) and the genomic 3′-BTE, and the in trans interaction between the genomic 3′ BTE and the 5′ UTR of SG RNA1 (Rakotondrafara et al., 2006). Numbers depict the nucleotide positions in the viral genomic RNA. The nucleotides participating in the long-range base-pairing are joined by lines.