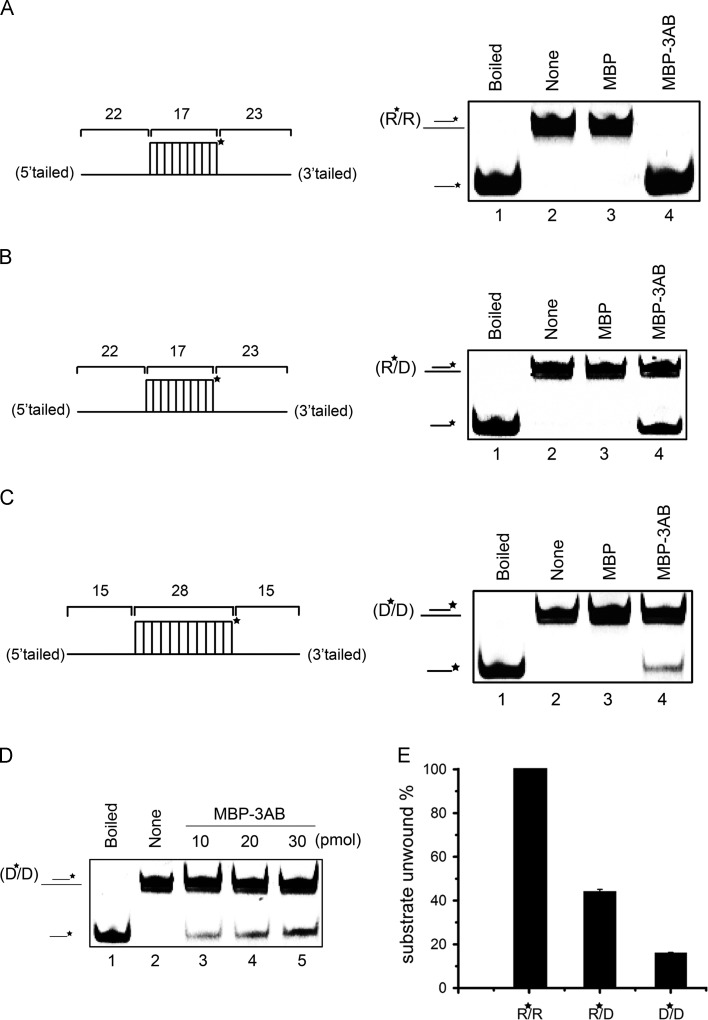
Fig. 2.
EV71 3AB can destabilize both RNA and DNA helices. (A–C) Purified MBP-3AB was incubated with standard RNA duplex (R*/R substrate), RNA/DNA hybrid duplex (R*/D substrate) and DNA duplex (D*/D substrate) as illustrated in left panels. Asterisk (*) indicates HEX-labeled strand. The preparations of destabilizing substrates are indicated in Materials and Methods. The 0.1 pmol substrate was incubated in standard reaction mixtures in the presence or absence of 10 pmol MBP-3AB as indicated, and the destabilizing activity was assessed via gel electrophoresis and scanning on a Typhoon 9200. (A) R*/R substrate (left panel). Lane 1, boiled reaction mixture without enzyme addition; Lane 2, reaction mixture without enzyme addition; Lane 3, complete reaction mixture with negative control MBP alone; Lane 4, complete reaction mixture with MBP-3AB. (B) R*/D substrate (left panel). Lanes 1 and 2: boiled (lane 1) or native (lane 2) reaction mixtures without enzyme addition; Lane 3, complete reaction mixture with negative control MBP alone; Lane 4, complete reaction mixture with MBP-3AB. (C) D*/D substrate (left panel). Lanes 1 and 2: boiled (lane 1) or native (lane 2) reaction mixtures without enzyme addition; Lane 3, complete reaction mixture with negative control MBP alone; Lane 4, complete reaction mixture with MBP-3AB. (D) Increasing amount of MBP-3AB was incubated with 0.1 pmol DNA duplex (D*/D) as indicated, and the destabilizing activity was assessed via gel electrophoresis and scanning on a Typhoon 9200. Asterisk (*) indicates HEX-labeled strand. (E) The helix destabilization was plotted as the percentage of the released RNA or DNA from the total helix substrates (Y-axis) at indicated helix substrates as X-axis.