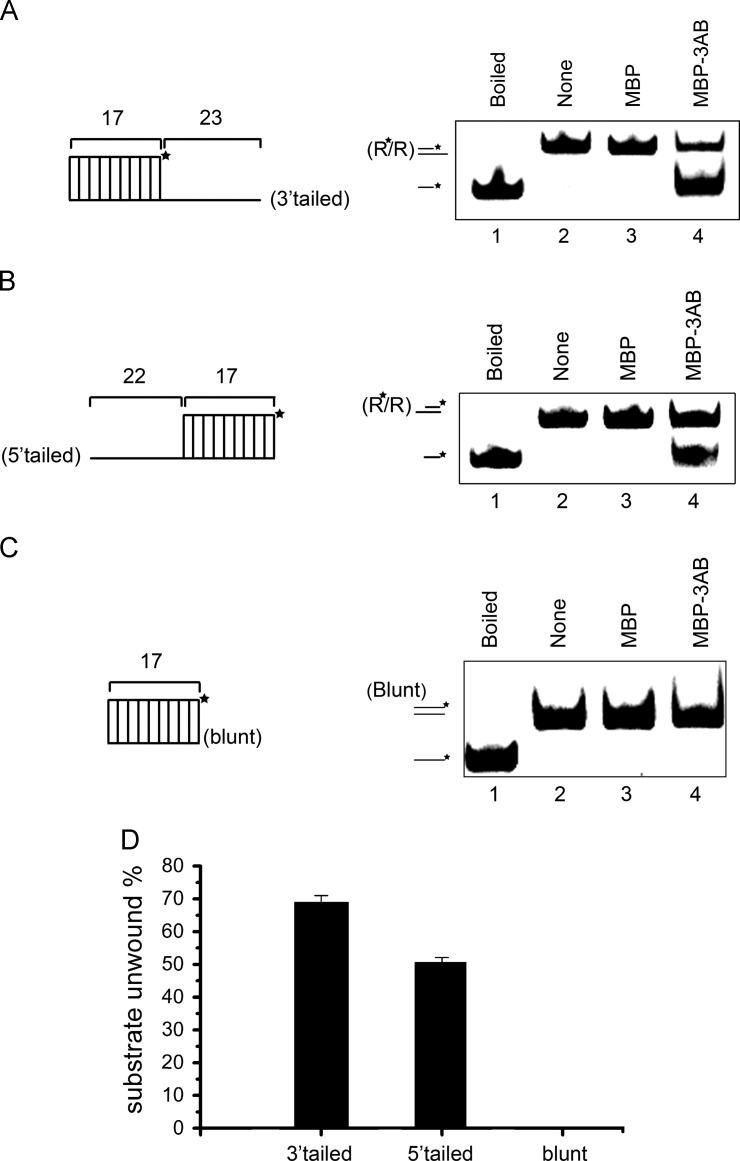
Fig. 3.
EV71 3AB destabilizes RNA helices in a bidirectional manner. (A) Negative control MBP alone (lane 3) and MBP-3AB (10 pmol) were reacted with 3′-tailed (lane 4) RNA helix substrate (0.1 pmol) as illustrated in upper panel, respectively, under standard reaction conditions. Asterisk (*) indicates HEX-labeled strand. (B) Negative control MBP alone (lane 3) and MBP-3AB (10 pmol) were reacted with 5′-tailed (lane 4) RNA helix substrate (0.1 pmol) as illustrated in upper panel, respectively, under standard reaction conditions. For panels A and B, Boiled 3′-tailed (A) or 5′-tailed (B) substrate (lane 1) or substrates alone (lanes 2) were used as controls. (C)MBP-3AB (10 pmol) was reacted with blunt-ended RNA helix (0.1 pmol) as illustrated in upper panel. (D) The unwinding activity was plotted as the percentage of the released RNA from the total RNA helix substrates (Y-axis) at indicated helix substrates as X-axis. Error bars represent standard deviation values from three separate experiments.