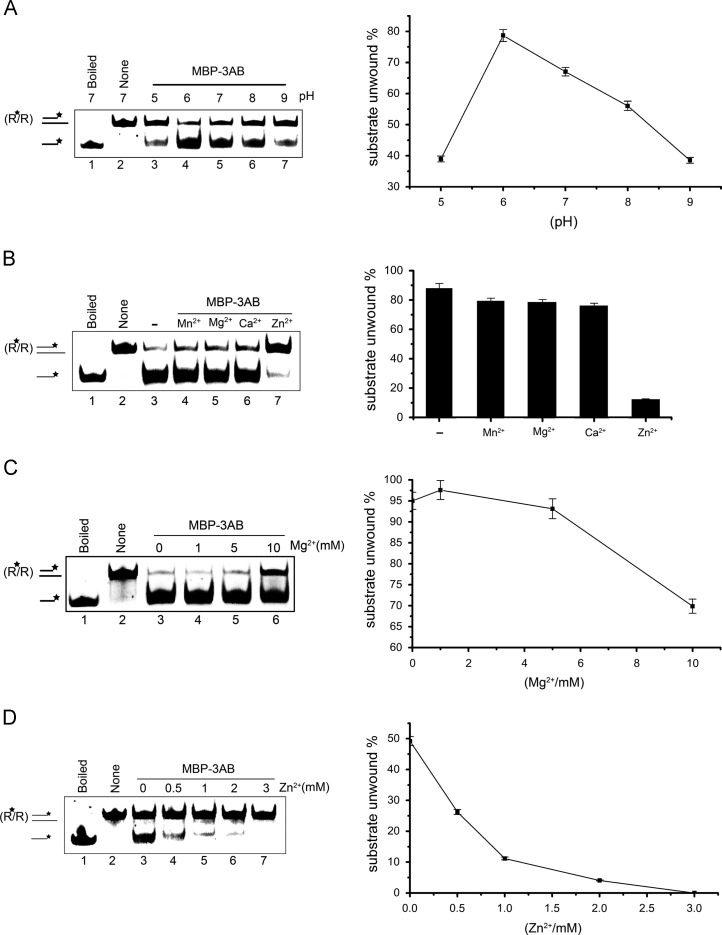
Fig. 5.
Biochemical conditions for the RNA helix-destabilizing activity of EV71 3AB. (A) Helix-destabilizing activity was determined at the indicated pH in the absence of divalent metal ions. (B) Standard RNA helix substrate (3′ tailed) (0.1 pmol) was reacted with MBP-3AB in the presence of the indicated divalent metal ions at 2 mM. (C and D) 3′-tailed RNA helix substrate (0.1 pmol) was reacted with MBP-3AB in the presence of indicated concentrations of MgCl2 (C) or ZnCl2 (D). Asterisks (*) indicate the HEX-labeled strand. For panels A–D, boiled and native substrates without MBP-3AB addition were used as controls (lane 1 and lane 2). For (A-D, right panels), the unwinding activity was plotted as the percentage of the released RNA from the total RNA helix substrates (Y-axis) at indicated pH values (A), divalent metal ions (B), Mg2+ concentrations (C), or Zn2+ concentration (D) as X-axis.