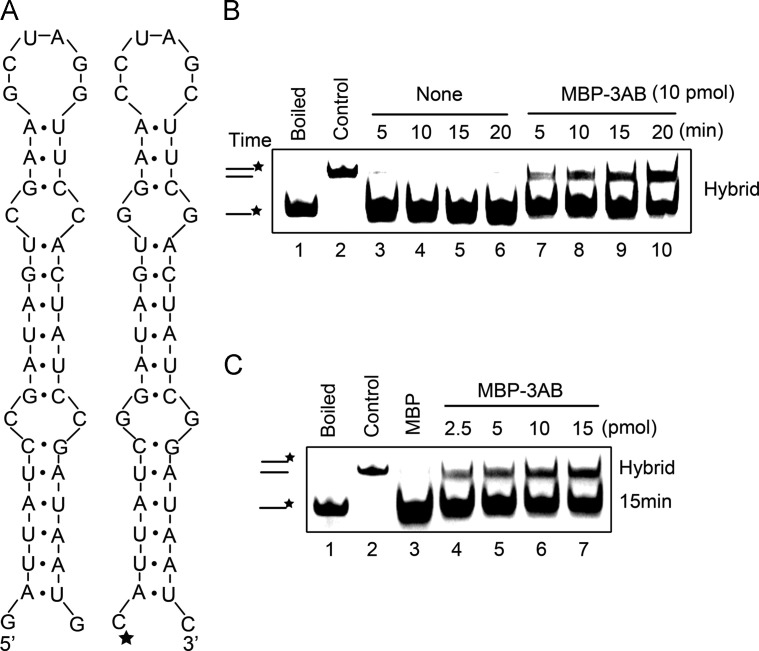
Fig. 6.
EV71 3AB destabilizes structured RNA strands. (A) Schematic illustrations of the predicted stem-loop structures of the two 42-nt RNA substrates. The two RNA strands are complementary. One strand has HEX-labeled 5′ end as indicated by asterisk (right), while the other strand was not labeled (left). (B) The two complementary strands were mixed (0.1 pmol each), and reacted in the absence (lanes 3–6) or presence (lanes 7–10) of MBP-3AB (10 pmol) for indicated time (5–20 min). (C) The two complementary strands were mixed (0.1 pmol each), and reacted with increasing amounts of MBP-3AB (2.5–15 pmol) for 15 min. Complete reaction mixture with negative control MBP alone (10 pmol) (lane 3). For (B-C), the mix of the two strands was boiled (to prevent spontaneous annealing) (lane 1) or treated with a thermal cycler (lane 2) as negative or positive control, respectively. Samples were subjected to gel electrophoresis (see Materials and methods).