Fig. 2.
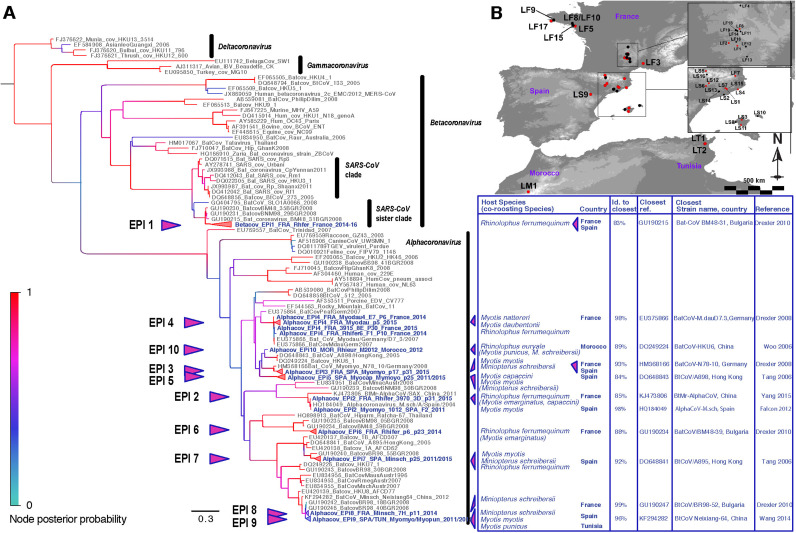
Phylogenetic analysis and geo-localisation of coronaviruses detected in the Western Palearctic region. A) Bayesian phylogeny depicted in Fig. 1 (selected nodes collapsed for clarity reasons) showing clusters including a number of sequences ranging from 3 to 63 with a mean of 14 per clade. Statistical support (posterior probability) of nodes are depicted using a gradual color code of the tree, red indicating significant posterior probability values (>0.95). Labels of viruses detected in this study are bolded and coloured in blue. Corresponding host names are indicated in the right panel, in front of each taxa reported in the study. When significant mixing of species at the roost was observed, the name of the co-roosting coronavirus-negative species is added in brackets. Country of origin and identity score (ID) to the closest reference found in GenBank were also added for each coronavirus clade detected in this study. B) Map of the study region depicting the 39 investigated sites and highlighting in red those where bat samples were found positive for coronaviruses.