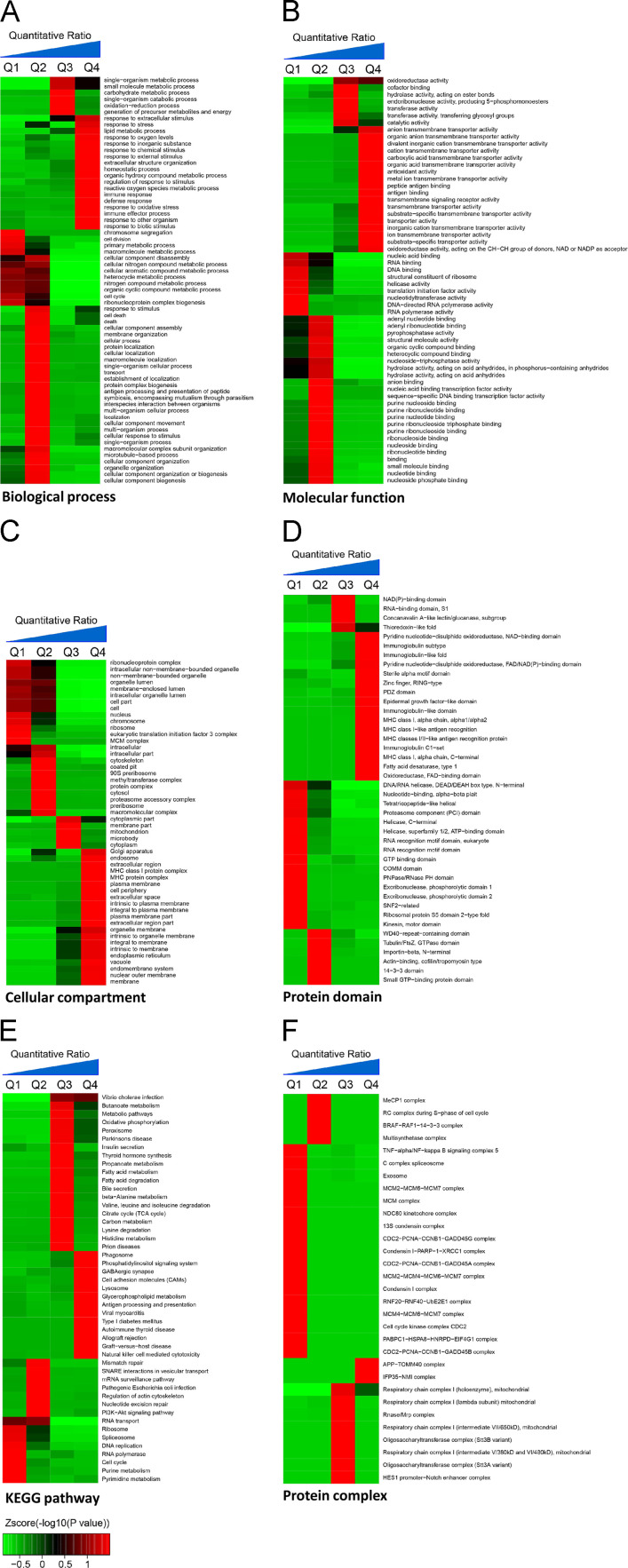
Fig. 2.
Enrichment and clustering analysis of the quantifiable proteomic data set. Quantifiable proteins were classified by gene ontology annotation based on (A) molecular function, (B) cellular compartment, and (C) biological process. In each category, the differential quantifiable proteins were divided into four quantiles based on the cumulative distribution of SILAC L/H ratios: Q1, less than 15%; Q2, 15–50%; Q3, 50–85%; and Q4, greater than 85%. An enrichment analysis was separately performed in each quantile for diverse categories, and the overrepresented annotations were clustered through one-way hierarchical clustering for comparative analysis. Quantifiable proteins were also annotated based on (D) the PFAM domain database, (E) the KEGG pathway database, and (F) the CORUM protein complex database.