Abstract
Cyclophilin A (CypA) is an important host factor in the replication of a variety of RNA viruses. Also the replication of several nidoviruses was reported to depend on CypA, although possibly not to the same extent. These prior studies are difficult to compare, since different nidoviruses, cell lines and experimental set-ups were used. Here, we investigated the CypA dependence of three distantly related nidoviruses that can all replicate in Huh7 cells: the arterivirus equine arteritis virus (EAV), the alphacoronavirus human coronavirus 229E (HCoV-229E), and the betacoronavirus Middle East respiratory syndrome coronavirus (MERS-CoV). The replication of these viruses was compared in the same parental Huh7 cells and in CypA-knockout Huh7 cells generated using CRISPR/Cas9-technology. CypA depletion reduced EAV yields by ~ 3-log, whereas MERS-CoV progeny titers were modestly reduced (3-fold) and HCoV-229E replication was unchanged. This study reveals that the replication of nidoviruses can differ strikingly in its dependence on cellular CypA.
Keywords: Cyclophilin, CypA, Arterivirus, MERS-coronavirus, EAV, human coronavirus-229E, CRISPR/Cas9, Knockout
Highlights
-
•
Nidoviruses display differences in sensitivity towards cyclophilin A depletion.
-
•
Replication of MERS-coronavirus is reduced modestly in cyclophilin A-knockout cells.
-
•
Equine arteritis virus replication is strongly inhibited by cyclophilin A depletion.
-
•
Chromosomal anomalies complicate CRISPR/Cas9-mediated gene editing in Huh7 cells.
1. Introduction
The order Nidovirales is currently comprised of the arterivirus, coronavirus, ronivirus, and mesonivirus families (https://talk.ictvonline.org/taxonomy/) and includes agents that can have major economic and societal impact. This was exemplified by the 2002–2003 severe acute respiratory syndrome coronavirus (SARS-CoV) epidemic and the ongoing outbreaks of the Middle East respiratory syndrome coronavirus (MERS-CoV). Both these coronaviruses were introduced into the human population following zoonotic transmission, revealing the potentially lethal consequences of nidovirus-induced disease in humans. Within a few months, the emergence of SARS-CoV led to more than 8000 laboratory-confirmed cases (mortality rate of ~ 10%). The MERS-CoV outbreaks thus far resulted in over 2000 confirmed human cases and a ~ 35% mortality rate within that group (http://www.who.int/emergencies/mers-cov/en/). In addition, the porcine epidemic diarrhea coronavirus and the arterivirus porcine reproductive and respiratory syndrome virus (PRRSV) are among the leading veterinary pathogens, having caused high economic losses in the swine industry (Holtkamp et al., 2013, Lin et al., 2016). The economic and societal impact of nidovirus infections, and the lack of effective strategies to control them, highlight the importance of advancing our knowledge of the replication of these viruses and their interactions with the host cell.
Nidoviruses are positive-stranded RNA (+RNA) viruses with large to very large genomes, ranging from 13 to 16 kb for arteriviruses to 26–34 kb for coronaviruses (Gorbalenya et al., 2006, Nga et al., 2011). Their complex genome expression strategy involves genome translation to produce the polyprotein precursors of the nonstructural proteins (nsps) as well as the synthesis of a nested set of subgenomic (sg) mRNAs to express the structural proteins (reviewed in de Wit et al., 2016, Snijder et al., 2013). Nidoviral nsps, presumably together with various host factors, assemble into replication and transcription complexes (RTCs) that drive viral RNA synthesis (Gosert et al., 2002, Hagemeijer et al., 2012, Pedersen et al., 1999, van Hemert et al., 2008a, van Hemert et al., 2008b). These RTCs are thought to be associated with a virus-induced network of endoplasmic reticulum (ER)-derived membrane structures, including large numbers of double-membrane vesicles (Gosert et al., 2002, Knoops et al., 2012, Knoops et al., 2008, Maier et al., 2013, Pedersen et al., 1999, Ulasli et al., 2010).
Nidovirus replication thus depends on a variety of host cell factors and processes, including cellular proteins and membranes, membrane trafficking, and host signaling pathways (reviewed in de Wilde et al., 2017b; van der Hoeven et al., 2016; Zhong et al., 2012). Among these, members of the cyclophilin (Cyp) protein family previously have been implicated in nidovirus replication. Cyclophilins are a family of peptidyl-prolyl isomerases (PPIases) that act as chaperones to facilitate protein folding, as well as protein trafficking and immune cell activation (reviewed in Naoumov, 2014; Nigro et al., 2013). Cyclophilins, and in particular the ubiquitously expressed CypA, have also been implicated in the replication of various other groups of RNA viruses. The role of CypA in hepatitis C virus (HCV) and human immunodeficiency virus-1 (HIV-1) infection has been studied in most detail. For example, CypA assists HCV polyprotein processing, interacts with HCV NS5A to ensure remodelling of cellular membranes into HCV replication organelles, and stabilizes HIV-1 capsids to promote nuclear import of the HIV-1 genome (reviewed in Hopkins and Gallay, 2015).
Cyclophilins were initially implicated as host factors in nidovirus replication during studies with general Cyp inhibitors such as cyclosporine A (CsA). In cell culture, the replication of a variety of coronaviruses and arteriviruses was found to be strongly inhibited by low-micromolar concentrations of CsA and the non-immunosuppressive CsA analogs Alisporivir (ALV) and NIM-811 (Carbajo-Lozoya et al., 2014, Carbajo-Lozoya et al., 2012, de Wilde et al., 2017a, de Wilde et al., 2013b, de Wilde et al., 2011, Kim and Lee, 2014, Tanaka et al., 2012, von Brunn, 2015, von Brunn et al., 2015). Subsequently, it was established that nidovirus replication can depend specifically on CypA and/or CypB. The replication in cell culture of the arterivirus equine arteritis virus (EAV; de Wilde et al., 2013a) and the alphacoronaviruses feline coronavirus (FCoV; Tanaka et al., 2017), human coronavirus (HCoV) NL63 (Carbajo-Lozoya et al., 2014), and HCoV-229E (von Brunn et al., 2015) was reported to be affected by CypA knockdown (KD) or knockout (KO), although the level of CypA dependence of these viruses, which was not compared directly, appeared to be quite different. Finally, upon ultracentrifugation, the normally cytosolic CypA was found to co-sediment with membrane structures containing EAV RTCs, suggesting a direct association with the arteriviral RNA-synthesizing machinery (de Wilde et al., 2013a).
The abovementioned studies differed in terms of the nidoviruses and cell lines tested, CypA expression levels, and readouts used to measure viral replication efficiency, which hampered a direct comparison of the CypA dependence of different nidoviruses. Therefore, in this study, we investigated the CypA-dependence of the replication of three distantly related nidoviruses in the same cell line (Huh7), in which CypA expression was knocked-out using CRISPR/Cas9 gene editing technology (Huh7-CypAKO cells). Using different cell lines, the replication of two of these viruses, the arterivirus EAV (de Wilde et al., 2013a) and the alphacoronavirus HCoV-229E (von Brunn et al., 2015), was previously concluded to depend on CypA. The CypA dependence of betacoronaviruses like MERS-CoV has not been documented before. Infection of Huh7-CypAKO cells with MERS-CoV revealed that its replication was only modestly affected by the absence of CypA, as opposed to EAV which was strongly inhibited. Strikingly, and in contrast to a previous report, HCoV-229E replication was not affected at all in the Huh7-CypAKO cells. Our study thus reveals major differences in the magnitude of CypA dependence of the arterivirus EAV when compared to two different coronaviruses, and calls for a more extensive evaluation of the role of CypA in the replication of members of the latter virus family.
2. Material and methods
2.1. Cell culture, infection, and virus titration
293T (van Kasteren et al., 2012), BHK-21 (Nedialkova et al., 2010), Huh7, and Vero cells (de Wilde et al., 2013b) were cultured as described previously. A cell culture-adapted derivative of the EAV Bucyrus isolate (Bryans et al., 1957, den Boon et al., 1991), HCoV-229E (ATCC VR-740; Hamre and Procknow, 1966), or MERS-CoV (strain EMC/2012; van Boheemen et al., 2012; Zaki et al., 2012) were used to infect monolayers of Huh7 cells (and derived cell clones) as described previously (Cervantes-Barragan et al., 2010, de Wilde et al., 2013b, Oudshoorn et al., 2016). EAV, HCoV-229E, and MERS-CoV titers in cell culture supernatants were determined by plaque assays on BHK-21, Huh7, or Vero cells, respectively (de Wilde et al., 2013b, Nedialkova et al., 2010, Zust et al., 2011).
2.2. Generation of cyclophilin knockout Huh7 cells
To obtain Huh7 cells that lack the expression of CypA, CypB, CypC or CypD, we used clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 gene editing (Cong et al., 2013). The pLentiCRISPR v2 plasmid (a kind gift from Feng Zhang; Addgene plasmid #52961; Sanjana et al., 2014) was used to construct plasmids with guide RNAs (gRNAs) specific for PPIA, PPIB, PPIC or PPID, or non-targeting gRNA (OriGene; for sequences, see Table 1), according to the protocol provided by Addgene (https://www.addgene.org/52961/). gRNA sequences were selected using the ATUM CRISPR gRNA design tool (https://www.atum.bio/eCommerce/cas9/input). Pseudo-infectious, third-generation lentivirus particles expressing gene-specific gRNAs were produced by transfection of 293 T cells with the pLentiCRISPR v2 plasmid and three “helper” plasmids (encoding HIV-1 gag–pol, HIV-1 rev, and VSV-G; Carlotti et al., 2004). After overnight transfection of 293T cells using the polyethyleneimine DNA co-precipitation method, the culture medium was replaced with fresh medium, which was harvested at 72 h post-transfection (h p.t.), passed through a filter with 0.45 µm pore-size, and stored at −80 °C. Huh7 cells were transduced in 10-cm2 dishes containing 1 ml of lentivirus harvest and 1 ml of fresh Huh7 culture medium supplemented with 8 µg/ml polybrene (Sigma). From 3 days post transduction onwards, lentivirus-transduced cells were selected by culturing in the presence of 3 µg/ml puromycin. Loss of cyclophilin expression was verified by Western blot analysis (see below).
Table 1.
gRNA sequences used in this study.
| Target | GeneIDa | Forward gRNAb | Reverse gRNA |
|---|---|---|---|
| Non-targeting | n.a. | CACCGGCACTACCAGAGCTAACTCA | AAACTGAGTTAGCTCTGGTAGTGCC |
| PPIA | 5478 | CACCGTGCCAGGACCCGTATGCTTT | AAACAAAGCATACGGGTCCTGGCAC |
| PPIB | 5479 | CACCGTTGCCGCCGCCCTCATCGCG | AAACCGCGATGAGGGCGGCGGCAAC |
| PPIC | 5480 | CACCGCTACCTCTCGTGCTTTGCGT | AAACACGCAAAGCACGAGAGGTAGC |
| PPID | 5481 | CACCGACTGAGAACCGTTTGTGTTG | AAACCAACACAAACGGTTCTCAGTC |
GeneID: a unique gene identifier that is assigned to a species-specific gene record in Entrez Gene (http://www.ncbi.nlm.nih.gov/gene).
gRNA, guide RNA.
To clone Huh7-CypAKO cells, lentivirus-transduced cells were trypsinized and seeded in 96-well culture plates at a density of one cell per well, which was verified by microscopy. Cell proliferation was stimulated by supplementing the culture medium with 15% FCS. Following their expansion, CypAKO clones were tested for lack of CypA expression by Western blot analysis.
The Huh7-CypAKO clones constitutively expressed the Cas9 nuclease as the gene is incorporated in the pLentiCRISPR v2 vector, and integrated into the genome upon lentivirus transduction. Using an alternative approach, we also generated of Huh7-CypAKO clones lacking Cas9 expression by direct transfection of the pLentiCRISPR v2 plasmid into Huh7 cells. Cells (5 × 105 cells in a 10-cm2 dish) were transfected with 2.5 µg plasmid DNA and 6 µl Lipofectamine 2000 (Thermo Fischer Scientific). After 24 h at 37 °C, the medium was replaced with fresh medium containing 3 µg/ml puromycin. At 4 d p.t., cells were harvested and seeded in 96-well culture plates at a density of one cell per well, to obtain clones as described above.
2.3. Antibodies and Western blot analysis
For Western blot analysis, we used rabbit polyclonal antibodies against CypA (sc-20360-R) or CypD (sc-66848) and a goat polyclonal antibody against CypB (sc-20361) (all from Santa Cruz Biotechnology), a rabbit monoclonal antibody against CypC (ab184552, Abcam), and mouse monoclonal antibodies against β-actin (AC-74, Sigma) or transferrin receptor (TfR; H68.4, Thermo Fisher Scientific). Cells were lysed in Laemmli sample buffer and after SDS-PAGE, proteins were transferred to Hybond-LFP membranes (GE Healthcare) by semi-dry blotting. Membranes were blocked with 1% casein in PBS containing 0.1% Tween-20 (PBST), and were incubated in PBST with 0.5% casein with anti-CypA (1:500), anti-CypB (1:1000), anti-CypC (1:1000), anti-CypD (1:500), anti-TfR (1:4000), or anti-β-actin (1:50,000) antisera. Biotin-conjugated swine-anti-rabbit IgG (1:2000) or goat-anti-mouse IgG (1:1000) antibodies (DAKO) and Cy3-conjugated mouse-anti-biotin (1:2500) were used for detection. Blots were scanned with a Typhoon 9410 imager (GE Healthcare) and analyzed with ImageQuant TL software.
2.4. Sequence analysis of Huh7-CypAKO clones
Genomic DNA was isolated from five different Huh7 CypAKO clones to verify the generation of Huh7 PPIA knockout cells. Approximately 3 × 106 cells were lysed in 3 ml of 75 mM NaCl, 25 mM EDTA pH 8.0, 1% SDS, and 100 µg/ml proteinase K, and incubated at 37 °C for 16 h. One ml of 6 M (saturated) NaCl was added and after mixing for 30 s, the lysate was centrifuged for 15 min at 4000×g and 4 °C. To remove remaining cellular debris, the supernatant was transferred to a new tube and centrifugation was repeated. DNA was precipitated in 70% EtOH, pelleted by centrifugation and washed in 70% EtOH. The dried pellet was dissolved in 10 mM Tris, 1 mM EDTA pH 7.5. For sequence analysis of the PPIA-gene, intron-specific primers (5′-ACCTTGCAGATTTGGCACAC-3′ and 5′-AGTGTTTGTTCCGTTCCCCC-3′) were used to amplify exon 4 of the PPIA gene that is targeted by CRISPR/Cas9-mediated cleavage (Fig. 2D). PCR products were cloned into pCR2.1-TOPO (Thermo Fisher Scientific) according to the manufacturer's instructions and individual clones were analyzed by Sanger sequencing to identify CRISPR/Cas9-directed out-of-frame insertions or deletions (‘indels’). In this manner, two Huh7 CypAKO clones (clones #1 and #2) were obtained, which were derived from lentiviral transduction and direct transfection of Huh7 cells, respectively.
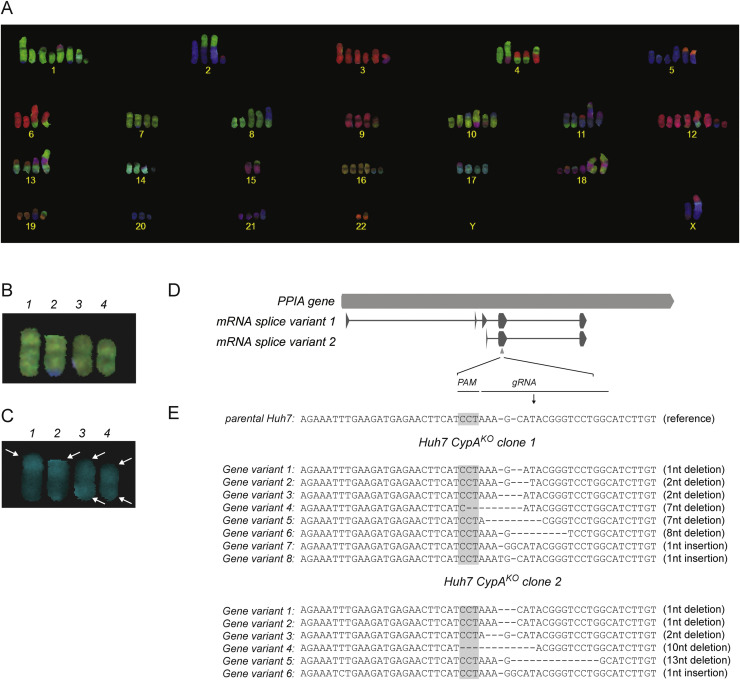
Fig. 2.
Karyotyping of Huh7 cells and sequencing of Huh7-CypAKOclones. A) Representative COBRA FISH karyogram of a Huh7 cell. A complex karyotype with both numerical and large number of structural rearrangements was observed. B) The short and long arm-specific whole chromosome paint probe set (for details, see Szuhai and Tanke, 2006) identified four copies of chromosome 7. C) Staining of the chromosomes’ short arms revealed the presence of two copies of isochromosome 7p (indicated as nr 3 and 4) and at least six copies of chromosome 7p (indicated by white arrows). D) Overview of the PPIA gene and PPIA-specific mRNA splice variants, including PPIA exons 1–5. The gRNA binding site is indicated by a grey triangle. E) Sequence analysis of cloned PCR products covering the PPIA-gene region targeted by the gRNA revealed eight (clone #1) or six (clone #2) out-of-frame mutations. Sequence analysis of clones derived from the parental Huh7 cells only yielded the reference PPIA gene sequence. The protospacer adjacent motif (PAM) sequence, which served as Cas9 docking site, is highlighted in grey.
2.5. Huh7 cell karyotyping
Chromosome spreads of metaphase cells were prepared essentially as described previously (de Graaff et al., 2015) and used for karyotyping by combined binary ratio labeling of nucleic-acid probes for multi-color fluorescence in situ hybridization (COBRA-FISH) staining. The analysis used whole chromosome paint probe sets to discriminate individual chromosomes and specific staining to identify short and long arms of chromosomes, as described previously (Szuhai and Tanke, 2006). After hybridization, image capture and image analysis was performed using in-house written software and chromosomes were identified based on their specific coloring (Szuhai and Tanke, 2006). Based on the chromosome count, ploidy (numerical aberrations) was established and structural chromosomal aberrations including breaks, deletions, translocations and insertions, were identified based on chromosomal structure.
2.6. Quantitative real-time RT-PCR analysis
Parental Huh7 cells and CypAKD Huh7 cells were cultured overnight in triplicate wells at a density of 5 × 104 cells in a 24-well cluster. Intracellular RNA was isolated using the Nucleospin RNA II kit (Machery-Nagel) according to the manufacturer's instructions and used as a template for cDNA synthesis using RevertAid H Minus Reverse Transcriptase (Thermo Fisher Scientific) and oligo(dT)20 primer. Finally, samples were assayed by quantitative real-time PCR analysis (qRT-PCR) using primers for PPIA (forward primer: TTCATCTGCACTGCCAAGAC; reverse primer: CAGACAAGGTCCCAAAGACAG), and the housekeeping genes RPL13 (forward primer: AAGGTGGTGGTCGTACGCTGTG; reverse primer: CGGGAAGGGTTGGTGTTCATCC) and GAPDH (forward primer: GCAAATTTCCATGGCACCGT; reverse primer: GCCCCACTTGATTTTGGAGG). PCR was performed using the IQ SYBR Green Super Mix (Bio-Rad) and the CFX384 Touch™ Real-Time PCR Detection System. Data was analyzed with CFX manager 3.1 software (Bio-Rad).
3. Results
3.1. Nidovirus replication in cyclophilin-knockout cell pools
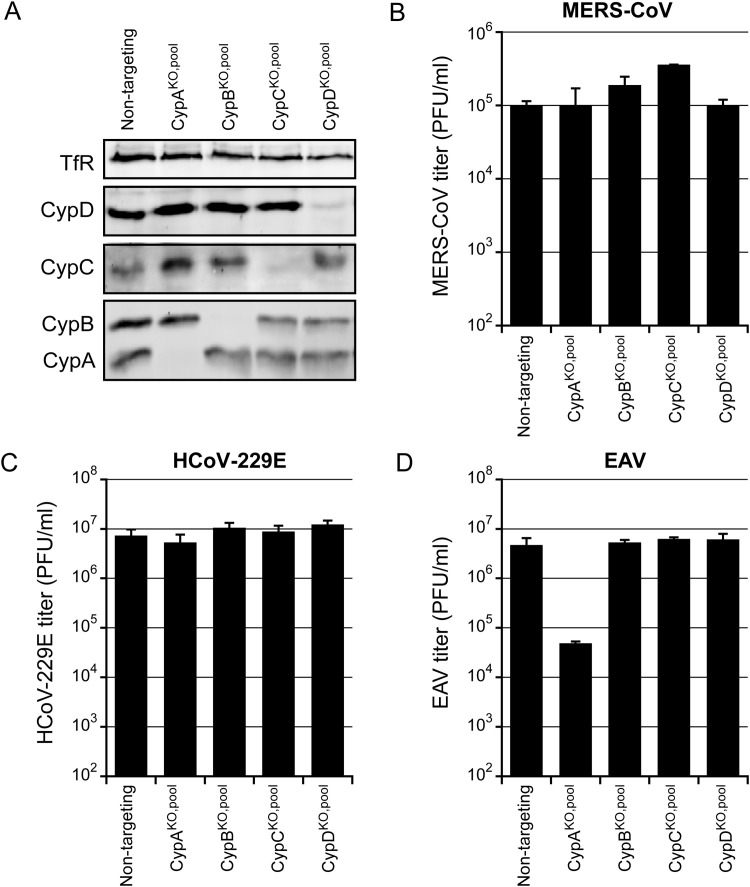
Previously, CypA and/or CypB were reported to play a role in the replication of a number of arteri- and coronaviruses (Carbajo-Lozoya et al., 2014, de Wilde et al., 2013a, Tanaka et al., 2017, von Brunn et al., 2015). The degree of sensitivity to Cyp depletion appeared to differ between these viruses, but was not compared directly. As we set out to assess the Cyp dependence of the recently emerged MERS-CoV, which had not been evaluated thus far, the fact that this betacoronavirus replicates in Huh7 cells provided an excellent opportunity for a direct comparison with the arterivirus EAV and the alphacoronavirus HCoV-229E, which can also replicate in this cell line. Therefore, using CRISPR/Cas9 gene editing technology, we first generated CypAKO, CypBKO, CypCKO, and CypDKO Huh7 cell pools. To this end, Huh7 cells were transduced with lentiviruses co-expressing the nuclease Cas9 as well as a gRNA that binds a specific sequence in the PPIA, PPIB, PPIC, or PPID gene. CypC and CypD were included given that both these enzymes are also sensitive to CsA treatment (Davis et al., 2010). The fact that it previously was implicated as a host factor in the replication of the betacoronavirus HCoV-OC43 (Favreau et al., 2012) was an additional reason to include CypD.
Western blot analysis of the CypKO cell pools established that CypA and CypB were no longer detectable in Huh7-CypAKO,pool and Huh7-CypBKO,pool cells, respectively. For the CypCKO or CypDKO cell pools, however, some residual expression of the target gene was detected ( Fig. 1A). Next, the replication of EAV, HCoV-229E, and MERS-CoV in each of the four CypKO cell pools was compared to their replication in Huh7 control cells expressing an unrelated (non-targeting) gRNA. To allow multiple cycles of replication before analyzing virus yields, cell pools were infected with a low infectious dose (MOI 0.01). The production of infectious MERS-CoV and HCoV-229E progeny was found to be unchanged in all four CypKO cell pools (Fig. 1B and C). In contrast, EAV virus titers were reduced by 2-logs after infection of the Huh7-CypAKO cell pool, but not in any of the other knockout cell pools (Fig. 1D). This strengthened the case for an important role of specifically CypA as a host factor in EAV replication. Previously, siRNA-mediated knockdown of CypA expression could only reduce EAV titers by about 4-fold (de Wilde et al., 2013a), suggesting that relatively small amounts of CypA may suffice to support normal levels of EAV replication.
Fig. 1.
Nidovirus replication in cyclophilin-knockout cell pools. A) Huh7 cells were transduced with lentiviruses expressing PPIA-, PPIB-, PPIC-, or PPID-specific gRNAs (Huh7-CypKO,pool cells) to achieve CRISPR/Cas9-mediated knockout of gene expression. The cell pools were analyzed for CypA, CypB, CypC, and CypD protein expression by Western blot analysis. Transferrin Receptor was used as loading control. B-D) Huh7-CypKO,pool cells were infected with MERS-CoV (B), HCoV-229E (C), or EAV (D) at an MOI of 0.01. Virus yields at 48 h p.i. (B, C) or 32 h p.i. (D) were determined by plaque assay. Results presented are the mean ± SD of triplicate harvests from a representative experiment.
3.2. Generation, characterizing, and karyotyping of Huh7-CypAKO clones
For HCoV-NL63, it was previously reported that virus production was inhibited only when the residual PPIA mRNA level was reduced to 3% of that in control cells (Carbajo-Lozoya et al., 2014). Also the results with EAV (see above) suggested that major effects on virus replication may only be observed when CypA knockdown is highly efficient. As the Huh7-CypAKO cell pools might exhibit low levels of residual CypA expression, which might still suffice to support normal levels of MERS-CoV and HCoV-229E replication, we generated clonal Huh7-CypAKO cell lines (as described in Material and Methods) for use in follow-up experiments.
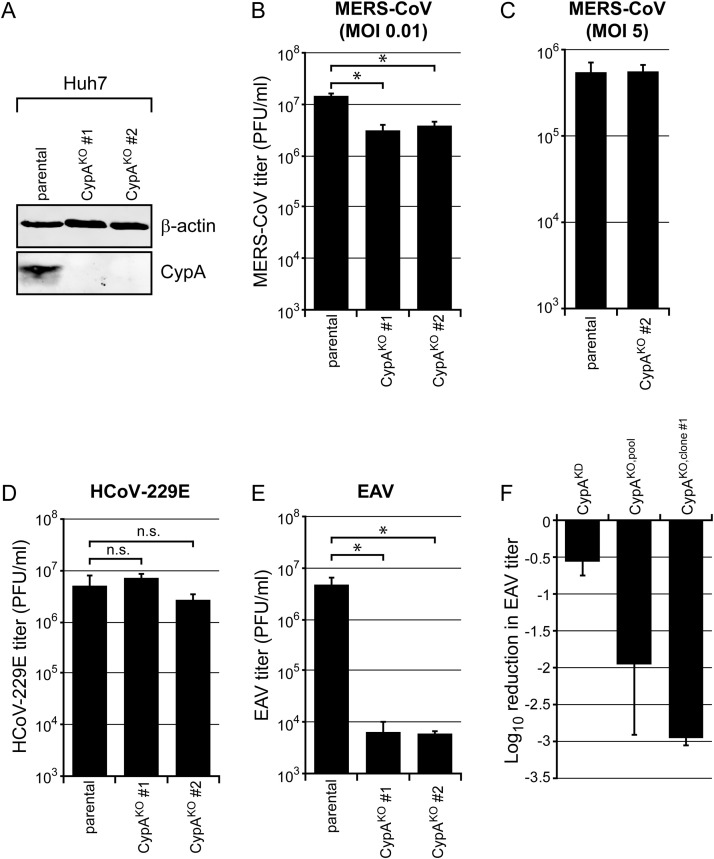
Potential knockout clones were first tested for CypA expression by Western blot analysis. Subsequently, the presence of out-of-frame ‘indels’ was established by sequence analysis of exon 4 of the PPIA gene. In total, five individual CypAKO cell clones were analyzed in detail. To this end, the exon that was targeted by the PPIA-specific gRNA was PCR amplified using primers targeting the adjacent introns ( Fig. 2D, white triangles) and the PCR product was cloned in a plasmid vector. Per CypAKO cell clone, over twenty plasmid clones were analyzed by Sanger sequencing. Remarkably, sequence analysis of these clones revealed six to eight different indel sequences (Fig. 2E; see also below), establishing that these Huh7 clones carried more than the anticipated two copies (alleles) of the PPIA gene. For three out of five cell clones, some of the indels constituted in-frame deletions, indicating that (low amounts of) CypA variants lacking one or more amino acids might still be expressed (data not shown). Two of the Huh7-CypAKO cell clones were found to contain only out-of-frame indels (Fig. 2E, clones #1 and #2), which was supported by a lack of detectable CypA expression ( Fig. 3A). Of note, one of these clones was obtained after lentivirus transduction (clone #1), whereas the other resulted from transfection of the pLentiCRISPR plasmid (clone #2).
Fig. 3.
Differences in EAV, MERS-CoV and HCoV-229E sensitivity to cyclophilin A depletion. A) Huh7-CypAKO cell clones #1 and #2 were analyzed for lack of CypA expression by Western blot analysis. Beta-actin was used as loading control. B, D-E) Parental Huh7 cells and Huh7-CypAKO clones #1 and #2 were infected with MERS-CoV (B), HCoV-229E (D), or EAV (E) at an MOI of 0.01. (C) Parental Huh7 cells and Huh7-CypAKO clone #2 were infected with MERS-CoV at an MOI of 5. Virus yields at 48 h p.i. (B,D), 24 h p.i. (C) or 32 h p.i. (E) were determined by plaque assay. Results are the mean ± SD of triplicate harvests from a representative experiment. F) Comparison of EAV progeny yields in Huh7-CypAKD (Chatterji et al., 2009), Huh7-CypAKO,pool cells (used in Fig. 1), and Huh7-CypAKO cell clone #1 (used in Fig. 3), presented as the difference in virus yields between Huh7 control cells and CypAKO or CypAKD cells (in log10 reduction in EAV titer). Results are the mean ± SD from two independent experiments.
To investigate the unexpectedly large number of different PPIA-gene specific sequences in more detail, we performed karyotyping of the parental Huh7 cells and Huh7-CypAKO clone #1. For both cell lines, COBRA-FISH established a 4n+ chromosome content, with more than 100 chromosomes per cell and exceedingly large numbers of translocations involving all chromosomes (results from the parental Huh7 cells are shown in Fig. 2A). The PPIA gene is located on the short arm of chromosome 7 (chr7p) and karyotyping using the short- and long arm-specific whole chromosome paint probe set (Szuhai and Tanke, 2006) revealed four copies of that chromosome, with one appearing normal (see chromosome 1; Fig. 2B), one carrying a translocation with a piece of chromosome 5 in the long arm (see chromosome 2; Fig. 2B), and two representing isochromosomes (i.e. containing two short arms of chromosome 7; see chromosomes 3 and 4; Fig. 2B). Staining of the short arm of chromosome 7 (chr7p) revealed six copies of this chromosome in the parental Huh7 cells (Fig. 2C, white arrows) and Huh7-CypAKO clone #1 (data not shown).
Overall, eight (clone #1) and six (clone #2) different PPIA-specific indels were identified. Based on the karyotyping analysis, this could be explained by the presence of (at least) six copies of the short arm of chromosome 7. As the resolution of this techniques is limited to the detection of large (>1000 kb) translocations (Szuhai and Tanke, 2006), an even higher number of PPIA gene copies in the Huh7 genome cannot be excluded, and would in fact be in line with the results obtained for clone #1. Since the analysis of >20 cloned PPIA-specific PCR products from CypAKO clones #1 and #2 exclusively revealed out-of-frame indels, both clones were assumed to have lost all expression of functional CypA and were used for further infection experiments.
3.3. Differences in EAV, MERS-CoV and HCoV-229E sensitivity to cyclophilin A-knockout
Parental Huh7 cells and Huh7-CypAKO clones #1 and #2 were all infected with MERS-CoV, HCoV-229E, or EAV at an MOI of 0.01. The inactivation of CypA expression in these cells was found to significantly reduce MERS-CoV replication by ~ 3-fold in both CypAKO cell clones (Fig. 3B). However, virus yields released from control cells and CypAKO cell clone #2 were similar upon high MOI inoculation (MOI of 5; Fig. 3C). Interestingly, in both clones, no effect of a lack of CypA was seen for HCoV-229E (Fig. 3D). For EAV, however, a ~ 3-log reduction of virus yields was observed (Fig. 3E), representing a 10-fold stronger inhibition compared to the Huh7-CypAKO,pool cells used previously (Fig. 1D).
4. Discussion
The importance of CypA in virus replication has been reported for a variety of viruses, including HCV and HIV-1. Also nidovirus replication has been shown to depend on CypA, but the role of this host factor is poorly understood thus far. By analogy with the role of CypA in HCV replication (reviewed in Hopkins and Gallay, 2015), and supported by the reduced replication of EAV in siRNA-treated CypAKD cells, we hypothesized CypA to be an integral component of the membrane-associated replication machinery of arteriviruses (de Wilde et al., 2013a; De Wilde et al., in preparation).
Previously, the role of CypA in nidovirus replication has been studied by multiple laboratories, with different outcomes. However, the use of a range of nidoviruses and cell lines, and variable experimental set-ups and read-outs likely contributed to these differences. By infecting the same Huh7-CypKO cell pools (Fig. 1) and Huh7-CypAKO clones (Fig. 3) with the three nidoviruses compared in this study, we could now eliminate a number of experimental variables. Our data revealed striking differences between the arterivirus EAV and the two coronaviruses in terms of their sensitivity to CypA-depletion in Huh7 cells. Whereas EAV replication was strongly inhibited (Fig. 3), coronavirus replication was affected only modestly (MERS-CoV) or not at all (HCoV-229E). Interestingly, the relatively small difference in MERS-CoV progeny titers from CypAKO and control Huh7 cells was not observed when infections were carried out with a high MOI (Fig. 3C). A possible explanation for this difference could be the presence of CypA in MERS-CoV virions. Virion-associated CypA has been reported previously for HIV-1 and SARS-CoV (Braaten et al., 1996, Hatziioannou et al., 2005, Neuman et al., 2008), and although its role and importance are unclear, the virion-mediated transfer of CypA to CypA-deficient cells may initially suffice to support efficient replication in these cells. Like SARS-CoV particles, MERS-CoV virions may contain CypA, which could explain complementation of the lack of CypA in knockout cells during a high-MOI, one-cycle replication assay. During a multi-cycle infection experiment, however, the virus produced during the first round of replication may lack CypA, which may negatively influence virus yields during subsequent rounds of replication.
In the case of HCoV-229E, the comparable replication efficiency in Huh7-CypAKO cells and control Huh7 cells appears to be at odds with published studies for three different alphacoronaviruses (including HCoV-229E itself) in which a modest to strong inhibition of virus replication was reported (see below). Likewise, the insensitivity (Fig. 1) of the alphacoronavirus HCoV-229E and betacoronavirus MERS-CoV to depletion of CypB and CypD, contrasts with previous reports on the CypB and CypD dependence of FCoV (Tanaka et al., 2017) and HCoV-OC43 (Favreau et al., 2012), an alpha- and a betacoronavirus respectively.
Significant efforts were made to characterize two of the Huh7-CypAKO clones that were generated using CRISPR/Cas9-technology: clone #1 was derived from transduction with a PPIA gRNA-expressing lentivirus, whereas clone #2 resulted from transfection of a plasmid co-expressing the PPIA-specific gRNA and the Cas9 nuclease. Consequently, clone #1, in contrast to clone #2, ubiquitously expressed Cas9 and the PPIA-gene directed gRNA. In theory, this could lead to off-target cleavage events in the genome, and thus to unwanted side-effects. However, using both clones, we observed very similar effects of CypA depletion on the replication of the three nidoviruses tested.
Previously documented differences in terms of sensitivity to CsA treatment appear to coincide with similar sensitivity differences to CypA knockdown as observed in this study. While EAV replication in cell culture is completely blocked at a dose of 1 µM CsA, concentrations of 9 and 16 µM are required to achieve a similar block of the replication of MERS-CoV (de Wilde et al., 2013b) and HCoV-229E (de Wilde et al., 2011, Pfefferle et al., 2011), respectively. An unexpected result was the lack of an effect of CypA depletion on HCoV-229E replication, since a previous study reported a 10-fold reduction of the HCoV-229E-driven expression of a luciferase reporter gene in Huh7.5-shRNA-CypAKD cells (von Brunn et al., 2015). In our hands, however, HCoV-229E replication was unchanged when comparing parental Huh7 cells, Huh7-CypAKO,pool cells, and the two Huh7-CypAKO clones (Figs. 1C and 3C). These contradictory results may (in part) be explained by the use of different viruses (wild-type versus recombinant HCoV-229E), the use of different readouts of virus replication (HCoV-229E progeny titers versus HCoV-229E-driven luciferase expression), or the use of Huh7 versus Huh7.5 cells. The latter is a sub-clone of Huh7 cells that supports HCV replication more efficiently, most likely because Huh7.5 cells express an inactive form of the retinoic acid-inducible gene-I (RIG-I), resulting in a reduced antiviral response to virus infection (Blight et al., 2002). Even more striking, is the difference with previous results obtained for two other alphacoronaviruses, FCoV and HCoV-NL63, for which progeny titer reductions of >5-fold and 5-log were reported upon CypA knockdown, respectively (Carbajo-Lozoya et al., 2014, Tanaka et al., 2017).
CypA is one of the most abundant cytosolic proteins constituting 0.1–0.4% of the total cellular protein content (Harding et al., 1986). Data from different labs suggest that relatively low levels of CypA may suffice to support efficient coronavirus replication. For example, we have previously shown that SARS-CoV replication was not affected when siRNA transfection was used to reduce the CypA expression level to 25% of that in parental cells (de Wilde et al., 2013a, de Wilde et al., 2011). HCoV-NL63 yields in Caco-2 cells were reduced only when a ~ 97% reduction in CypA mRNA expression was achieved (Carbajo-Lozoya et al., 2014). Subsequently, FCoV replication was found to be completely abolished in CRISPR/Cas9-mediated CypAKO fcwf-4 (feline) cells, while only partial inhibition of FIPV replication was observed when CypA knockdown was mediated by short-hairpin (sh)RNAs (Tanaka et al., 2017).
We observed a similar correlation between remaining CypA expression levels and EAV replication. This study was initiated using Huh7 cells in which CypA knockdown was mediated by constitutive expression of shRNAs (Fig. 3E; Chatterji et al., 2009). In these cells, CypA protein expression was undetectable and the CypA mRNA level was only 2% of that in control Huh7 cells expressing a non-targeting shRNA (data not shown). In these cells, EAV replication was reduced by about 0.5-log (Fig. 3F, left bar). Also in our Huh7-CypAKO cell pools (Figs. 1D and 3F) residual CypA expression could not be detected by Western blot analysis, but now a ~ 2-log reduction in EAV yield was observed. The stronger inhibition of EAV suggests that CypA levels in these cells were lower than those in shRNA-expressing CypAKD cells, but due to the nature of the gene editing method used (targeting the PPIA gene instead of the mRNA) CypA depletion could not be readily compared by measuring mRNA levels. Finally, the largest reductions of EAV progeny yields, by about ~ 3-logs, were observed using our Huh7-CypAKO cell clones (Fig. 3B-F).
Interestingly, the characterization of our Huh7-CypAKO clones and the parental Huh7 cell line revealed a severely disordered chromosome composition with multiple translocations and duplications involving all chromosomes, including chromosome 7 that contains the PPIA gene. Such structural rearrangements, leading to gene expression abnormalities, are common in tumor-derived cell lines (Lin et al., 2014, Mertens et al., 2015), but are not always taken into account during studies of virus-host interactions. Since Huh7 cells originated from a hepatocellular carcinoma (Nakabayashi et al., 1982), these abnormalities were to be expected, also on the basis of a previous study that established chromosomal imbalances in liver carcinoma-derived cell lines, including Huh7 (Wilkens et al., 2012). Interestingly, this study describes a gain of, among other regions, the part of short arm of chromosome 7 that includes the PPIA gene. Polyploidy and chromosome rearrangements may hamper the generation of bona fide knockout cell lines. Therefore, our observations underline the importance of an in-depth genomic characterization of knockout cell lines obtained by (CRISPR/Cas9-mediated) gene editing. This is further emphasized by the relatively low success rate of our efforts to obtain true CypAKO cell clones, since more gene editing events are needed to achieve complete knockout. Several of our other clones were found to carry in-frame deletions in PPIA and (potentially) expressed CypA variants lacking one or multiple amino acid residues, with unpredictable consequences for the protein's function.
The replication of MERS-CoV, HCoV-229E, and EAV in CypBKO, CypCKO, or CypDKO cell pools was unaltered compared to control cells, suggesting that in the Huh7 setting these viruses do not depend on the presence of CypB, CypC, or CypD. However, as in the case of CypA, we cannot exclude the possibility that (very) low levels of these Cyps may still suffice to support efficient virus replication. This may be exemplified by the dependence of FCoV on CypB. Whereas FCoV replication in feline CypBKO fcwf-4 cells was strongly reduced, replication in shRNA-mediated CypBKD fcwf-4 cells was only marginally affected, suggesting that a very strong reduction of CypB levels is needed to block replication efficiently (Tanaka et al., 2017). Unfortunately, in the present study, we were unable to generate true Huh7-CypBKO, Huh7-CypCKO, and Huh7-CypDKO cell clones, but the analysis of nidovirus replication in the context of cloned cells lacking these Cyps definitely warrants further efforts. Likewise the quantitative and functional aspects of the role of CypA in nidovirus replication needs to be followed up, particularly in the light of the quite different results that can apparently be obtained with the same coronavirus in different experimental settings.
Acknowledgements
We thank Diede Oudshoorn for helpful discussions and technical assistance on creating CRISPR/Cas9-mediated Cyp knockout cells, Dr. Ron Fouchier (Erasmus Medical Center Rotterdam, The Netherlands) for sharing MERS-CoV isolate EMC/2012, and Dr. Volker Thiel for providing HCoV-229E.
Contributor Information
Adriaan H. de Wilde, Email: A.H.de_Wilde@lumc.nl.
Eric J. Snijder, Email: E.J.Snijder@lumc.nl.
References
- Blight K.J., McKeating J.A., Rice C.M. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braaten D., Franke E.K., Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryans J.T., Crowe M.E., Doll E.R., McCollum W.H. Isolation of a filterable agent causing arteritis of horses and abortion by mares; its differentiation from the equine abortion (influenza) virus. Cornell Vet. 1957;47:3–41. [PubMed] [Google Scholar]
- Carbajo-Lozoya J., Ma-Lauer Y., Malesevic M., Theuerkorn M., Kahlert V., Prell E., von Brunn B., Muth D., Baumert T.F., Drosten C., Fischer G., von Brunn A. Human coronavirus NL63 replication is cyclophilin A-dependent and inhibited by non-immunosuppressive cyclosporine A-derivatives including Alisporivir. Virus Res. 2014;184C:44–53. doi: 10.1016/j.virusres.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbajo-Lozoya J., Muller M.A., Kallies S., Thiel V., Drosten C., von Brunn A. Replication of human coronaviruses SARS-CoV, HCoV-NL63 and HCoV-229E is inhibited by the drug FK506. Virus Res. 2012;165:112–117. doi: 10.1016/j.virusres.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlotti F., Bazuine M., Kekarainen T., Seppen J., Pognonec P., Maassen J.A., Hoeben R.C. Lentiviral vectors efficiently transduce quiescent mature 3T3-L1 adipocytes. Mol. Ther.: J. Am. Soc. Gene Ther. 2004;9:209–217. doi: 10.1016/j.ymthe.2003.11.021. [DOI] [PubMed] [Google Scholar]
- Cervantes-Barragan L., Zust R., Maier R., Sierro S., Janda J., Levy F., Speiser D., Romero P., Rohrlich P.S., Ludewig B., Thiel V. Dendritic cell-specific antigen delivery by coronavirus vaccine vectors induces long-lasting protective antiviral and antitumor immunity. MBio. 2010;1:e00171–00110. doi: 10.1128/mBio.00171-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji U., Bobardt M., Selvarajah S., Yang F., Tang H., Sakamoto N., Vuagniaux G., Parkinson T., Gallay P. The isomerase active site of cyclophilin A is critical for hepatitis C virus replication. J. Biol. Chem. 2009;284:16998–17005. doi: 10.1074/jbc.M109.007625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T.L., Walker J.R., Campagna-Slater V., Finerty P.J., Paramanathan R., Bernstein G., MacKenzie F., Tempel W., Ouyang H., Lee W.H., Eisenmesser E.Z., Dhe-Paganon S. Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases. PLoS Biol. 2010;8:e1000439. doi: 10.1371/journal.pbio.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaff M.A., de Jong D., Briaire-de Bruijn I.H., Hogendoorn P.C., Bovee J.V., Szuhai K. A translocation t(6;14) in two cases of leiomyosarcoma: Molecular cytogenetic and array-based comparative genomic hybridization characterization. Cancer Genet. 2015;208:537–544. doi: 10.1016/j.cancergen.2015.07.005. [DOI] [PubMed] [Google Scholar]
- de Wilde A.H., Falzarano D., Zevenhoven-Dobbe J.C., Beugeling C., Fett C., Martellaro C., Posthuma C.C., Feldmann H., Perlman S., Snijder E.J. Alisporivir inhibits MERS- and SARS-coronavirus replication in cell culture, but not SARS-coronavirus infection in a mouse model. Virus Res. 2017;228:7–13. doi: 10.1016/j.virusres.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Li Y., van der Meer Y., Vuagniaux G., Lysek R., Fang Y., Snijder E.J., van Hemert M.J. Cyclophilin inhibitors block arterivirus replication by interfering with viral RNA synthesis. J. Virol. 2013;87:1454–1464. doi: 10.1128/JVI.02078-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Raj V.S., Oudshoorn D., Bestebroer T.M., van Nieuwkoop S., Limpens R.W., Posthuma C.C., van der Meer Y., Barcena M., Haagmans B.L., Snijder E.J., van den Hoogen B.G. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-alpha treatment. J. Gen. Virol. 2013;94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Snijder E.J., Kikkert M., van Hemert M.J. Host factors in coronavirus replication. Curr. Top. Microbiol. Immunol. 2017 doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Zevenhoven-Dobbe J.C., van der Meer Y., Thiel V., Narayanan K., Makino S., Snijder E.J., van Hemert M.J. Cyclosporin A inhibits the replication of diverse coronaviruses. J. Gen. Virol. 2011;92:2542–2548. doi: 10.1099/vir.0.034983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J.A., Snijder E.J., Chirnside E.D., de Vries A.A., Horzinek M.C., Spaan W.J. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favreau D.J., Meessen-Pinard M., Desforges M., Talbot P.J. Human coronavirus-induced neuronal programmed cell death is cyclophilin d dependent and potentially caspase dispensable. J. Virol. 2012;86:81–93. doi: 10.1128/JVI.06062-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 2002;76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Vonk A.M., Monastyrska I., Rottier P.J., de Haan C.A. Visualizing coronavirus RNA synthesis in time by using click chemistry. J. Virol. 2012;86:5808–5816. doi: 10.1128/JVI.07207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proceedings of the Society for Experimental biology and medicine. Soc. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- Harding M.W., Handschumacher R.E., Speicher D.W. Isolation and amino acid sequence of cyclophilin. J. Biol. Chem. 1986;261:8547–8555. [PubMed] [Google Scholar]
- Hatziioannou T., Perez-Caballero D., Cowan S., Bieniasz P.D. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J. Virol. 2005;79:176–183. doi: 10.1128/JVI.79.1.176-183.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkamp D.J., Kliebenstein J.B., Neumann E.J. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. JSHAP. 2013;21:72–84. [Google Scholar]
- Hopkins S., Gallay P.A. The role of immunophilins in viral infection. Biochim. Biophys. Acta. 2015;1850:2103–2110. doi: 10.1016/j.bbagen.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lee C. Porcine epidemic diarrhea virus induces caspase-independent apoptosis through activation of mitochondrial apoptosis-inducing factor. Virology. 2014;460–461:180–193. doi: 10.1016/j.virol.2014.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Barcena M., Limpens R.W., Koster A.J., Mommaas A.M., Snijder E.J. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J. Virol. 2012;86:2474–2487. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., van den Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Saif L.J., Marthaler D., Wang Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016;226:20–39. doi: 10.1016/j.virusres.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.C., Boone M., Meuris L., Lemmens I., Van Roy N., Soete A., Reumers J., Moisse M., Plaisance S., Drmanac R., Chen J., Speleman F., Lambrechts D., Van de Peer Y., Tavernier J., Callewaert N. Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat. Commun. 2014;5:4767. doi: 10.1038/ncomms5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H.J., Hawes P.C., Cottam E.M., Mantell J., Verkade P., Monaghan P., Wileman T., Britton P. Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. MBio. 2013;4:e00801–e00813. doi: 10.1128/mBio.00801-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens F., Johansson B., Fioretos T., Mitelman F. The emerging complexity of gene fusions in cancer. Nat. Rev. Cancer. 2015;15:371–381. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- Nakabayashi H., Taketa K., Miyano K., Yamane T., Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- Naoumov N.V. Cyclophilin inhibition as potential therapy for liver diseases. J. Hepatol. 2014;61:1166–1174. doi: 10.1016/j.jhep.2014.07.008. [DOI] [PubMed] [Google Scholar]
- Nedialkova D.D., Gorbalenya A.E., Snijder E.J. Arterivirus Nsp1 modulates the accumulation of minus-strand templates to control the relative abundance of viral mRNAs. PLoS Pathog. 2010;6:e1000772. doi: 10.1371/journal.ppat.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates J.R., 3rd, Wuthrich K., Stevens R.C., Buchmeier M.J., Kuhn P. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 2008;82:5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nga P.T., Parquet Mdel C., Lauber C., Parida M., Nabeshima T., Yu F., Thuy N.T., Inoue S., Ito T., Okamoto K., Ichinose A., Snijder E.J., Morita K., Gorbalenya A.E. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 2011;7:e1002215. doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro P., Pompilio G., Capogrossi M.C. Cyclophilin A: a key player for human disease. Cell Death Dis. 2013;4:e888. doi: 10.1038/cddis.2013.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudshoorn D., van der Hoeven B., Limpens R.W., Beugeling C., Snijder E.J., Barcena M., Kikkert M. Antiviral innate immune response interferes with the formation of replication-associated membrane structures induced by a positive-strand RNA virus. MBio. 2016:7. doi: 10.1128/mBio.01991-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K.W., van der M.Y., Roos N., Snijder E.J. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 1999;73:2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle S., Schopf J., Kogl M., Friedel C.C., Muller M.A., Carbajo-Lozoya J., Stellberger T., von Dall'armi E., Herzog P., Kallies S., Niemeyer D., Ditt V., Kuri T., Zust R., Pumpor K., Hilgenfeld R., Schwarz F., Zimmer R., Steffen I., Weber F., Thiel V., Herrler G., Thiel H.J., Schwegmann-Wessels C., Pohlmann S., Haas J., Drosten C., von Brunn A. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011;7:e1002331. doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Kikkert M., Fang Y. Arterivirus molecular biology and pathogenesis. J. Gen. Virol. 2013;94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- Szuhai K., Tanke H.J. COBRA: combined binary ratio labeling of nucleic-acid probes for multi-color fluorescence in situ hybridization karyotyping. Nat. Protoc. 2006;1:264–275. doi: 10.1038/nprot.2006.41. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Sato Y., Osawa S., Inoue M., Tanaka S., Sasaki T. Suppression of feline coronavirus replication in vitro by cyclosporin A. Vet. Res. 2012;43:41. doi: 10.1186/1297-9716-43-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Sato Y., Sasaki T. Feline coronavirus replication is affected by both cyclophilin A and cyclophilin B. J. Gen. Virol. 2017;98:190–200. doi: 10.1099/jgv.0.000663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulasli M., Verheije M.H., de Haan C.A., Reggiori F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 2010;12:844–861. doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D., Haagmans B.L., Gorbalenya A.E., Snijder E.J., Fouchier R.A. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3:e00473–00412. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoeven B., Oudshoorn D., Koster A.J., Snijder E.J., Kikkert M., Barcena M. Biogenesis and architecture of arterivirus replication organelles. Virus Res. 2016;220:70–90. doi: 10.1016/j.virusres.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert M.J., de Wilde A.H., Gorbalenya A.E., Snijder E.J. The in vitro RNA synthesizing activity of the isolated arterivirus replication/transcription complex is dependent on a host factor. J. Biol. Chem. 2008;283:16525–16536. doi: 10.1074/jbc.M708136200. [DOI] [PubMed] [Google Scholar]
- van Hemert M.J., van den Worm S.H., Knoops K., Mommaas A.M., Gorbalenya A.E., Snijder E.J. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4:e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kasteren P.B., Beugeling C., Ninaber D.K., Frias-Staheli N., van Boheemen S., Garcia-Sastre A., Snijder E.J., Kikkert M. Arterivirus and nairovirus ovarian tumor domain-containing Deubiquitinases target activated RIG-I to control innate immune signaling. J. Virol. 2012;86:773–785. doi: 10.1128/JVI.06277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Brunn A. Editorial overview: engineering for viral resistance. Curr. Opin. Virol. 14. 2015 doi: 10.1016/j.coviro.2015.08.009. (v-vii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Brunn A., Ciesek S., von Brunn B., Carbajo-Lozoya J. Genetic deficiency and polymorphisms of cyclophilin A reveal its essential role for Human Coronavirus 229E replication. Curr. Opin. Virol. 2015;14:56–61. doi: 10.1016/j.coviro.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkens L., Hammer C., Glombitza S., Muller D.E. Hepatocellular and cholangiolar carcinoma-derived cell lines reveal distinct sets of chromosomal imbalances. Pathobiology. 2012;79:115–126. doi: 10.1159/000334100. [DOI] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhong Y., Tan Y.W., Liu D.X. Recent progress in studies of arterivirus- and coronavirus-host interactions. Viruses. 2012;4:980–1010. doi: 10.3390/v4060980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S., Siddell S.G., Ludewig B., Thiel V. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]