Abstract
RNA replication of positive-strand (+)RNA viruses requires the lipids present in intracellular membranes, the sites of which viral replicases associate with. However, the direct effects of membrane lipids on viral replicases are still poorly understood. Wuhan nodavirus (WhNV) protein A, which associates with mitochondrial membranes, is the sole replicase required for RNA replication. Here, we report that WhNV protein A binds to RNA1 in a cooperative manner. Moreover, mitochondrial membrane lipids (MMLs) stimulated the RNA binding activity and cooperativity of protein A, and such stimulations exhibited strong selectivity for distinct phospholipids. Interestingly, MMLs stimulated the RNA-binding cooperativity only at higher protein A concentrations. Further investigation showed that MMLs stimulate the RNA binding of protein A by promoting its self-interaction. Finally, manipulating MML metabolism affected the protein A-induced RNA1 recruitment in cells. Together, our findings reveal the direct effects of membrane lipids on the RNA binding activity of a nodaviral replicase.
Keywords: Wuhan Nodavirus, Protein A, RNA binding, Mitochondrial membrane lipids
Highlights
-
•
WhNV protein A directly binds to RNA1 in a cooperative manner.
-
•
Mitochondrial membrane lipids (MMLs) stimulate the binding activity of protein A.
-
•
The RNA binding of protein A is selectively stimulated by specific phospholipids.
-
•
MMLs enhance the RNA binding of protein A by stimulating its self-interaction.
-
•
Manipulating phospholipid metabolism regulates protein A-induced RNA1 recruitment.
Introduction
(+)RNA viruses replicate their genomic RNA in viral RNA replication complexes (vRCs) including viral replicase proteins, viral RNA, and host proteins, on specific rearranged intracellular membranes (Ahlquist, 2006, Ahlquist et al., 2003, Miller and Krijnse-Locker, 2008, Sasvari and Nagy, 2010). Different viruses form their vRCs on diverse intracellular organelle membranes, including the endoplasmic reticulum (Diaz et al., 2012, Lee et al., 2001, Mas and Beachy, 1999, Pedersen et al., 1999, Schlegel et al., 1996, Schmidt-Mende et al., 2001), Golgi apparatus (Schlegel et al., 1996), lysosomes (Froshauer et al., 1988, Kujala et al., 2001, Magliano et al., 1998, Schlegel et al., 1996), endosomes (Froshauer et al., 1988, Kujala et al., 2001), peroxisomes (Jonczyk et al., 2007, White and Nagy, 2004) and mitochondria (Miller and Ahlquist, 2002, Miller et al., 2001). Lipids are major components of intracellular membranes, as they control membrane fluidity and plasticity (Nohturfft and Zhang, 2009, van Meer et al., 2008), and membrane lipids facilitate the RNA replication of (+)RNA viruses (Miller and Krijnse-Locker, 2008). For instance, the complete activity of Semliki Forest virus (SFV) NSP1 protein, an mRNA capping enzyme, requires association with specific negative phospholipids (Ahola et al., 1999). Sphingomyelin has been found to activate the RNA-dependent RNA polymerase (RdRp) activity of Hepatitis C virus (HCV) genotype 1b (Weng et al., 2010). The precise functions of intracellular membranes, particularly membrane lipids, in the RNA replication of (+)RNA viruses have not been fully understood, but possibly include offering an optimal microenvironment for viral replicase enzymatic activities, facilitating the use of membrane-associated host cofactors and/or directly interacting with viral replicases to mediate their functions (Chukkapalli et al., 2012, Heaton and Randall, 2011).
Nodaviruses (family Nodaviridae) are (+)RNA viruses that contain a bipartite genome consisting of RNA1 (3.1 kb) and RNA2 (1.4 kb), which encodes protein A, the RdRP (Gallagher et al., 1983), and capsid precursor protein ɑ (Schneemann et al., 1992), respectively. Moreover, a subgenomic RNA3 (sgRNA3), is synthesized during RNA1 replication and encodes protein B2, a suppressor of antiviral RNA interference (RNAi) (Li et al., 2002). Nodaviruses express a sole RNA replicase, protein A, which is responsible for both membrane association and viral RNA replication (Ball, 1995, Kopek et al., 2007, Miller and Ahlquist, 2002, Miller et al., 2001, Venter and Schneemann, 2008). This characteristic renders nodaviruses like Flock House virus (FHV) and WhNV simplified and ideal models for studying viral RNA replication.
In the case of FHV, the most extensively studied Nodaviridae member, its protein A is localized to outer mitochondrial membranes (Miller and Ahlquist, 2002, Miller et al., 2001). Previous studies of FHV indicated that membrane lipids mediate FHV RNA protein A function. The in vitro study showed that complete replication activity of FHV vRCs isolated from intracellular membrane require the addition of exogenous phospholipids (Wu et al., 1992, Wu and Kaesberg, 1991). FHV RNA replication in Drosophila cells can be blocked by inhibiting fatty acid synthesis (Kampmueller and Miller, 2005). Moreover, FHV protein A is a lipid-binding protein with particular affinity for specific anionic phospholipids, which may regulate the protein A-membrane interactions (Stapleford et al., 2009). However, although membrane lipids play an important role in nodaviral RNA replication, the detailed mechanisms by which membrane lipids regulate the activities of nodaviral protein A are not well understood.
As a virus closely related to FHV, WhNV has been well characterized and provides novel insights for nodaviral subgenomic RNA replication (Qiu et al., 2011), RNA silencing suppression (Qi et al., 2011, Qi et al., 2012) and initiation of RNA synthesis (Wang et al., 2013). Similar to FHV, WhNV protein A is associated with mitochondrial membranes (Qiu et al., 2013), and moreover, the membrane-association of WhNV protein A is closely linked with the recruitment and stabilization of viral genomic RNA1 on vRCs (Qiu et al., 2013). Furthermore, the in vitro data showed that WhNV protein A is self-interacted and the self-interaction of WhNV protein A is directly mediated by MMLs, suggesting the direct role of membrane lipids in WhNV protein A function (Qiu et al., 2014).
In this study, we focused on the direct effects of MMLs on the RNA-binding activity of WhNV protein A, because viral genomic RNA binding to viral replicases is important during many steps of the RNA replication, including the recruitment of the viral genomic RNA to the vRCs, recognition of replicating-elements, encapsidation of genomic RNA, assembly of the vRCs and activation of RdRp (Pathak et al., 2011, Pathak et al., 2012). To this end, we first examined whether WhNV protein A binds to RNA1 and RNA2 in vitro, and then explored the direct effects of MMLs on the RNA binding of protein A. We found that WhNV protein A binds to RNA1 in a cooperative manner, and the cooperativity could be stimulated by high protein A concentrations. MMLs stimulated the RNA-binding activity of protein A, and this stimulation exhibited strong selectivity for different phospholipids. Interestingly, MMLs stimulated the RNA-binding cooperativity only at higher but not at lower protein A concentrations. Further investigation showed that MMLs stimulate the RNA binding activity of protein A by promoting its self-interaction. Finally, we further confirmed that the MML composition changes by manipulating phospholipid metabolism affect protein A-induced RNA1 recruitment in cells.
Results
Characterization of the RNA probe used for determining the RNA binding of WhNV protein A
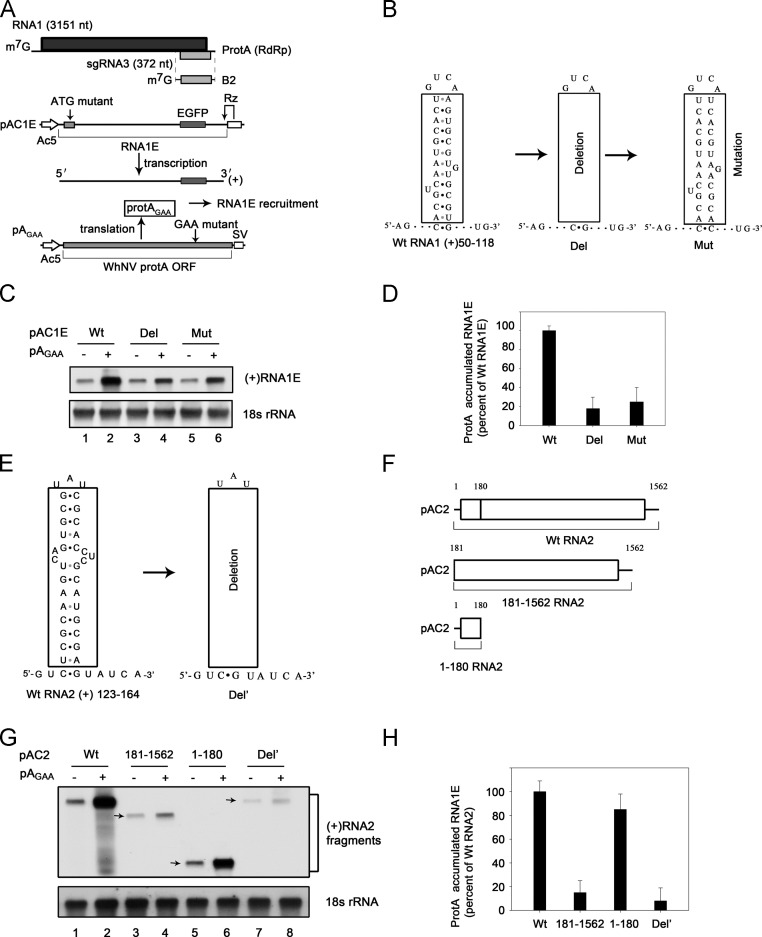
To determine whether WhNV protein A could bind to RNA directly, we first determined which RNA sequences can be used as the RNA probe. Previous study reported that nodaviruses contain the conversed sequences in the 5′-proximal region of RNA1 that exhibit a stem-loop structure and are responsible for RNA recruitment (Van Wynsberghe and Ahlquist, 2009). According to WhNV, its RNA1 nt 50–118 [RNA1(50–118)] exhibited a similar secondary structure ( Fig. 1B). To examine whether these sequences are responsible for WhNV RNA1 recruitment/stabilization in cells, we constructed two RNA1 derivates and examined their recruitment in the presence of WhNV replication-incompetent protein AGAA (Fig. 1A). The plasmid pAC1E, in which an (enhanced green fluorescent protein) EGFP open reading frame (ORF) is inserted at the 3′ end of RNA1 sequence (Qiu et al., 2013), is a functional template for RNA1 replication (the transcribed and replicated products are labeled as “RNA1E”), but the ORF of protein A is closed by the mutation of the start codon (Qiu et al., 2013). WhNV Protein AGAA is provided by the plasmid pAGAA. The RdRp activity of Protein AGAA is nulled by mutating the GDD replication site to GAA as previously described (Qiu et al., 2013, Wang et al., 2013). Protein AGAA retained its RNA1 recruitment activity despite losing its RNA replicase activity (Qiu et al., 2013).
Fig. 1.
Characterization of the RNA probe used for determining the RNA binding of WhNV protein A in vitro. (A) Schematic of plasmids used for protein AGAA (prot AGAA) and (+)RNA1E expression. RNA1E templates with authentic viral 5′ and 3′ termini of WhNV RNA1 and an inserting EGFP sequence were generated from pAC1E by precisely placing the Ac5 promoter start site and a hepatitisδribozyme (Rz), respectively, and by mutating the start codon at the indicated location to disrupt translation. The Ac5 promoter and SV40 polyadenylation signal (SV) flanking the protein A ORF in pAGAA thereof disrupt its activity as a viral RNA replication template and mutating the replication GDD sites into GAA but maintain its activity to recruit RNA (Qiu et al., 2014). pAGAA-derived protein AGAA subsequently directs (+)RNA1E recruitment from (+)RNA1E template transcribed from pAC1E. (B) The secondary structure predicted for RNA1 nt 50–118 [RNA1(50–118)]. Del represents removing the RNA sequences formed the helices structure and Mut represents destroying the base pairing in the helices section. (C) RNA1(50–118) mediates RNA1 recruitment in cells. Pr-E cells were transfected with the indicated plasmids, including pAC1E wt, Del or Mut (as shown in B) in the absence or in the presence of pAGAA (protein AGAA). After transfection for 36 h, total RNA was extracted and analyzed by Northern blot with the probes against EGFP and 18S rRNA, respectively. (D) The levels of (+)RNA1E were determined from three experiments after normalization to 18S rRNA and are expressed as the level of protein A-stimulated (+)RNA1E accumulation relative to wt (+)RNA1E. (E) The secondary structure predicted for RNA2 nt 123–164 [RNA1(123–164)]. Del′ represents removing the RNA sequences formed the helices structure. (F) Schematic of plasmids used for protein A-mediated RNA2 recruitment. (G) RNA2(123–164) mediates RNA2 recruitment in cells. Pr-E cells were transfected with the indicated plasmids, including pAC2 wt, pAC2 181–1562, pAC2 1–180, pAC2 Del′ (as shown in E and F) in the absence or in the presence of pAGAA. After transfection for 36 h, total RNA was extracted and analyzed by Northern blot with the probes against EGFP and 18S rRNA, respectively. (H) The levels of (+)RNA2 were determined from three experiments after normalization to 18S rRNA and are expressed as the level of protein A-stimulated (+)RNA2 accumulation relative to wt (+)RNA2.
We determined the sequences required for RNA1 recruitment in Pr-E cells, which are derived from Pieris rapae larvae, the natural host of WhNV (Qiu et al., 2014). Thirty-six hrs after transfecting with the pAGAA and the indicated plasmids, cells were collected and total RNA was analyzed by Northern blotting. As shown in Fig. 1C, the presence of protein AGAA supported the recruitment of wild type (wt) RNA1E (compared lane 2 to lane 1; Fig. 1D). While destroying the stem-loop by either deleting (Fig. 1B, “Del”) or mutating the stem structure (Fig. 1B, “Mut”) of RNA1E resulted in the decrease in RNA1E recruitment (Fig. 1C, compared lane 4 or 6 to lane 2; Fig. 1D). These results indicate that WhNV RNA1 nt 50–118 is needed for RNA1 recruitment.
We sought to determine the sequences required for RNA2 recruitment in cells. The analysis of RNA2 sequence using RNA mfold showed that RNA2 nt 123–164 [RNA2(123–164)] exhibits a stable stem-loop structure (Fig. 1E). We then examined whether this sequence is required for RNA2 recruitment by protein A in cells. Deletion of RNA2 nt 1–180 (Fig. 1F) resulted in a significant reduction in RNA2 recruitment in the presence of protein AGAA (Fig. 1G, compared lane 4 to lane 2; Fig. 1H). On the other hand, RNA2 nt 1–180 was sufficient to support protein A-mediated RNA2 recruitment (Fig. 1G, compared lane 6 to lane 5; Fig. 1H). Furthermore, deleting the stem-loop region (Fig. 1E) resulted in a significant decrease in RNA2 recruitment (Fig. 1G, comparing lane 8 to lane 2; Fig. 1H). These results indicate that WhNV RNA2 nt 123–164 is required for RNA2 recruitment. Together, both RNA1(50–118) and RNA2(123–164) can be used as the RNA probes for the gel mobility shift experiments.
The characterization of the RNA1 binding behavior of protein A revealed its cooperative RNA binding
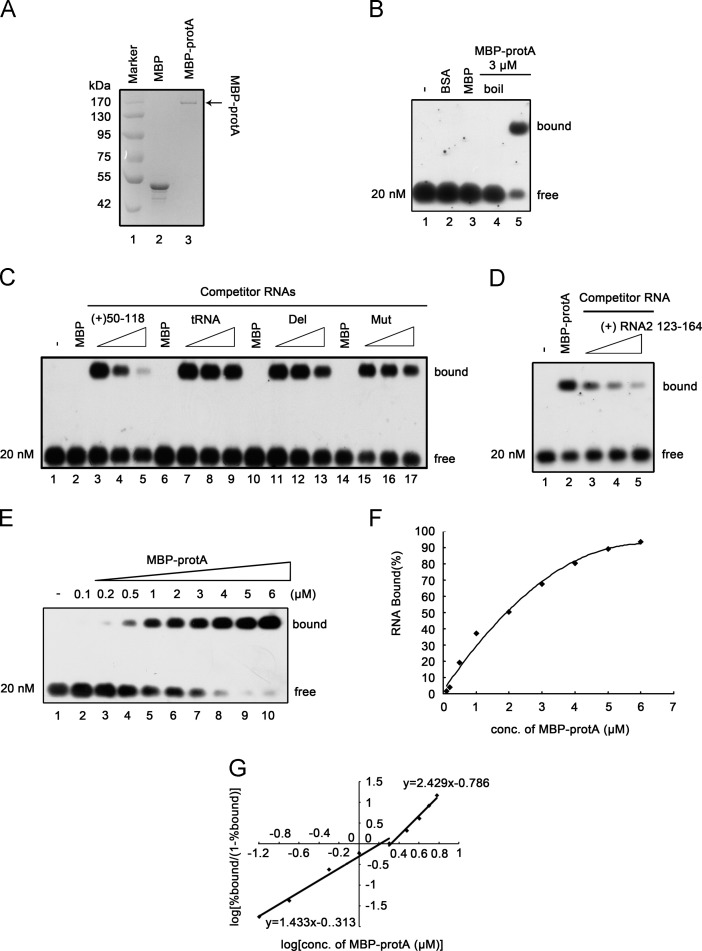
Maltose-binding protein (MBP)-tagged full-length (FL) protein A (MBP-protA) was expressed in Escherichia coli and purified (Qiu et al., 2014) (Fig. 1A). Standard gel mobility shift experiments with a DIG-labeled RNA1(50–118) were performed to determine the RNA binding ability of MBP-protA at 27 °C, and the samples were subject to 1.0% agarose gel electrophoresis. As shown in Fig. 2B, MBP-protA did bind to RNA efficiently (lane 5), whereas the negative controls (MBP alone, bovine serum albumin [BSA], and boiled MBP-protA) exhibited no RNA binding ability (lanes 2–4).
Fig. 2.
The binding preference of recombinant protein A to RNA1. (A) SDS-PAGE analysis of purified recombinant protein A from E. coli. Protein A ORF was cloned into pMAL-c2X and expressed as C-terminal fusion proteins with MBP (MBP-protA) as described previously (Qiu et al., 2014). Lane 1, Marker; lane 2, MBP protein alone; lane 3, MBP-protA. (B) Gel mobility shift assay showing interactions between MBP-protA and RNA1. The in vitro transcribed DIG-labeled RNA1(50–118) was separately incubated with bovine serum albumin (BSA, lane 2), MBP alone (lane 3), boiled MBP-protA (lane 4) and MBP-protA (lane 5) (3 μM each), in a binding buffer at 27 °C for 30 min and then analyzed in 1% agarose gel. Gel was transferred to Hybond N nylon membranes via capillary transfer and then the membranes were incubated with anti-DIG antibody conjugated with alkaline phosphatase, exposed to film. The unbound, free RNA1(50–118) probe and the shift (bound) RNA-protein complex are marked on the right. (C) Unlabeled competitor RNAs at increasing concentrations (in 1-, 10-, 60-fold excess) were added to the mixture containing the DIG-labeled RNA1(50–118) and 3 μM MBP-protA, and the bound complexes were analyzed in a gel mobility shift assay. The tRNA was from yeast. (D) Gel mobility shift assay showing interactions between MBP-protA and RNA2. The in vitro transcribed DIG-labeled RNA2(123–164) was incubated with MBP-protA (lane 2) and in a binding buffer at 27 °C for 30 min and then analyzed in 1% agarose gel. Unlabeled competitor RNAs at increasing concentrations (in 5-, 50-, 100-fold excess) were added to the mixture containing the DIG-labeled RNA2(123–164) and 3 μM MBP-protA, and the bound complexes were analyzed in a gel mobility shift assay. (E) Cooperative binding of MBP-protA to RNA1(50–118). Gel mobility shift assays were performed using increasing molar concentrations of MBP-protA incubated with 20nM RNA1(50–118) probe. The molar concentrations of MBP-protA (0.1–6 μM) are indicated above each lane. (F) The plot of the percent of RNA bound versus molar concentration of MBP-protA. (G) The Hill coefficients of the RNA binding of protein A based on Fig. 2E at low and high protein concentrations are indicated.
Moreover, RNA probe competition assays were performed to test whether MBP-protA specifically bound to RNA1(50–118) (Fig. 2C). Briefly, the same amounts of DIG-labeled RNA probe and MBP-protA were used in the presence of increasing amounts of unlabeled competitors, such as RNA1(50–118) wt, Del or Mut, and yeast tRNA (Fig. 2C). Only RNA1(50–118) wt efficiently competed with DIG-labeled RNA1(50–118), showing that protein A binds to RNA1 specifically. We also revealed that protein A can directly bind to RNA2 by the gel mobility shift experiment in the presence of MBP-protA and DIG-labeled RNA2(123–164) (Fig. 2D, lane 2). Moreover, increasing the amount of unlabeled RNA2(123–164) competitor resulted in the gradual reduction of the bound RNA probe, indicating that RNA2 is specifically bound to protein A as does RNA1 (Fig. 2D, lanes 3–5).
We used RNA1(50–118) as the RNA probe for the subsequent assays. To further characterize the behavior of protein A when it binds to RNA1, we incubated progressively increasing amounts of MBP-protA (0.1–6 μM) with 20 nM of DIG-labeled RNA1(50–118) (Fig. 2E). In the presence of a small amount of MBP-protA, the RNA probe was found mostly in free form, while increasing the amount of MBP-protA resulted in a rapid transition of the RNA probe to bound form (Fig. 2E). The minimal ratio of protein A to RNA probe for RNA binding is about 10:1 (Fig. 2E, lane 3; 0.2 μM of MBP-protA to 20 nM of RNA probe), while the bound band was clearly visible at the protein concentration of 0.5 μM (Fig. 2E, lane 4). The binding curve based on Fig. 2E also indicated that the amount of bound RNA is linked to the concentration of MBP-protA (Fig. 2F). And the apparent dissociation constant Kd was calculated as 1.9 μM for protein A at an RNA probe concentration of 20 nM (Fig. 2F). Moreover, the RNA binding data were quantified, and a Hill transformation was applied to determine whether the binding of protein A to RNA1 is cooperative [i.e., that the binding of protein A to RNA increased the affinity of protein A to bind to additional RNA molecules; a >1 Hill coefficient indicates positive cooperativity (Rajendran and Nagy, 2003)]. Interestingly, protein A at low concentrations (0.1–2 μM) had a Hill coefficient value of ~1.433, whereas high protein concentrations had a higher value of ~2.429 (Fig. 2G). These results indicate that protein A binds to RNA1 in a cooperative manner, and higher protein A concentrations (>2 μM) yield higher cooperativity.
MMLs stimulate the binding of WhNV protein A to RNA1
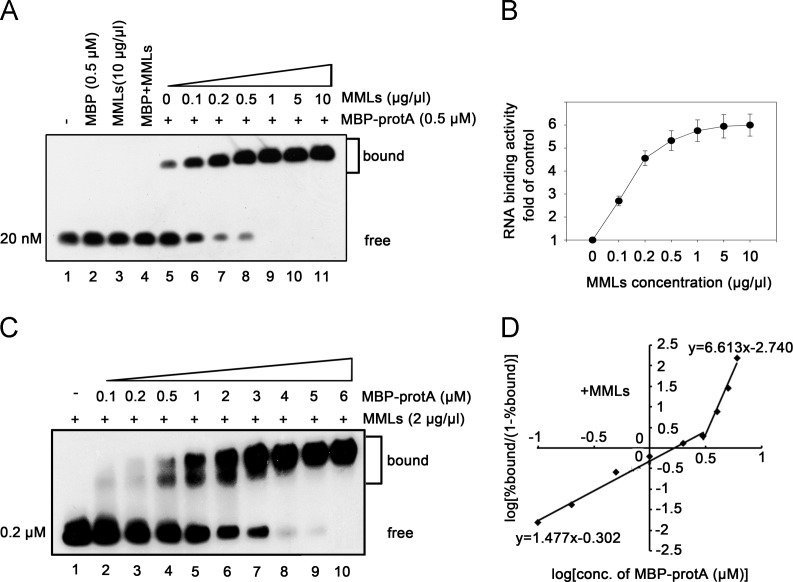
After determining that protein A directly binds to RNA, we sought to examine the direct effects of MMLs on the binding of WhNV protein A to RNA1. To this end, we isolated MMLs from the mitochondrial outer membranes of Pr-E cells as described in Materials and Methods. Since the 0.5 μM protein concentration is efficient for MBP-protA binding to RNA1 (Fig. 2E, lane 4), we performed the gel mobility shift experiments using 0.5 μM of MBP-protA with the addition of the increasing concentrations of MMLs ( Fig. 3A). The results showed that MMLs stimulated the RNA1 binding activity of WhNV protein A in a dose-response manner (Fig. 3A, lanes 5–11), whereas MMLs alone or in combination with negative control MBP protein did not bind to RNA1 (Fig. 3A, lanes 3 and 4). The RNA1 binding activity of protein A was enhanced about 2.7-fold at an MML concentration of 0.1 μg/μl, about 5.5-fold at an MML concentration of 1 μg/μl, and then plateaued at MML concentrations of 5 and 10 μg/μl (Fig. 3B).
Fig. 3.
MMLs stimulate the RNA binding activity of protein A. (A,B) MMLs stimulate the binding of protein A to RNA1(50–118) probe. Gel mobility shift assays were performed using increasing concentrations (wt/vol) of MMLs with 0.5 μM MBP-protA and 20 nM RNA1(50–118) probe. The concentrations of MMLs are indicated above each lane. The RNA binding activity of protein A in the absence of MMLs is used as the control (1-fold). The increase in the RNA binding activity of protein A at each point concentration of MMLs is graphed as the fold of control as shown in (B). Error bars represent S.D. values from at least three independent experiments. (C,D) MMLs stimulate protein A cooperatively binding to RNA1(50–118) at high protein concentrations. Gel mobility shift assays were performed using increasing molar concentrations of MBP-protA incubated with 0.2 μM RNA1(50–118) probe with the addition of 2 μg/μl MMLs, and then analyzed via 2% agarose gel to clearly separate migration of protein–RNA complexes. The Hill coefficients of the RNA binding of protein A in the presence of MMLs at low and high protein concentrations are indicated (D).
Furthermore, we sought to determine whether MMLs affect the cooperative binding of protein A to RNA1. To this end, we conducted RNA binding experiments under the conditions described in Fig. 2E except the addition of 2 μg/μl MMLs to the reaction mixtures and increasing the amount of RNA probe to 0.2 μM, because the activity of protein A binding to RNA is significantly stimulated in the presence of MMLs. Because the changes of the migration of protein–RNA complex with the addition of MMLs were not significant in 1.0% agarose gel (Fig. 2E), we used 2.0% agarose gel to separate the samples in this assay. As shown in Fig. 3C, increasing the amount of MMLs resulted in a rapid transition of the RNA probe to bound form and obvious changes of the migration of protein–RNA complex at the high protein concentrations. The RNA binding data was applied to Hill transformation (Fig. 3D) and compared to that in the absence of MMLs (Fig. 2G). Very interestingly, although the presence of MMLs had little effect on the cooperativity of protein A binding to RNA1 (Hill coefficient value: 1.477 with MMLs vs. 1.433 without MMLs) at lower concentrations of MBP-protA, it substantially stimulated the cooperativity at higher protein concentrations (>3 μM), as the Hill coefficient value was dramatically increased from 2.429 (Fig. 2G) to 6.163 (Fig. 3D). Taken together, we conclude that the cooperativity of protein A binding to RNA is stimulated by MMLs only at high protein A concentrations.
Specific anionic phospholipids stimulate WhNV protein A binding to RNA
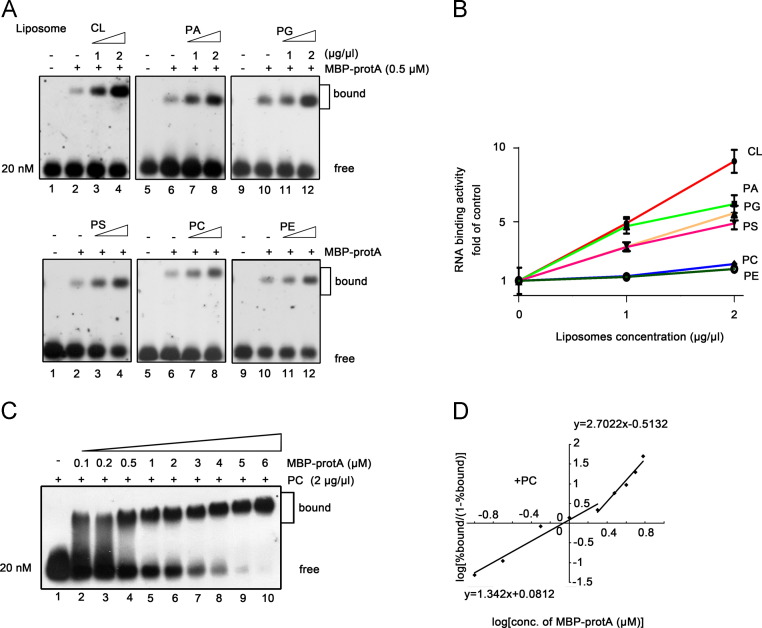
MMLs are composed of various specific phospholipids (van Meer et al., 2008). The various phospholipid compositions of intracellular membranes are the key determinants of the activities of membranes as well as membrane-associated proteins (van Meer et al., 2008). Thus, we further analyzed the in vitro RNA binding activity of protein A with liposomes that form mitochondrial outer membranes. These liposomes were generated from equal amounts of purified individual cellular phospholipids as described in Materials and Methods. A series of dose-response assays were performed to determine the effect of distinct liposomes on the RNA binding activity of protein A ( Fig. 4A), and the data was graphed as the fold of the RNA binding activity of protein A without lipids. As shown in Fig. 4B, anionic phospholipid 1,1′,2,2′-tetraoleoyl cardiolipin (CL) had substantial stimulation effects; anionic phospholipids 1,2-dioleoylsn-glycero-3-phosphate (PA), 1,2-dioleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (PG), and 1,2-dioleoylsn-glycero-3-[phospho-l-serine] (PS) had moderate stimulation effects; and zwitterionic lipids 1,2-dioleoyl-sn-glycero-3-phosphocholine (PC) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (PE) had minimal effects. These results show that RNA binding of protein A is selectively enhanced by specific anionic phospholipids.
Fig. 4.
Specific anionic phospholipids stimulate the RNA binding activity of protein A. (A,B) Gel mobility shift assays were performed using the increasing concentrations (wt/vol) of liposomes generated from specific phospholipids with 0.5 μM MBP-protA and 0.2 μM RNA1(50–118) probe complexes. The concentrations of liposomes are indicated above each lane. The RNA binding activity of protein A in the absence of liposomes is used as the control (1-fold). The increases in the RNA binding activity of protein A at each point concentration of indicated liposomes are graphed as the fold of control (B). Error bars represent S.D. values from at least three independent experiments. (C,D) PC showed limited effect on protein A cooperatively binding to RNA1(50–118). Gel mobility shift assays were performed using increasing molar concentrations of MBP-protA incubated with 20 nM RNA1(50–118) probe with the addition of 2 μg/μl PC. The Hill coefficients of the RNA binding of protein A in the presence of PC at low and high protein concentrations are indicated (D).
Since zwitterionic lipids showed no obvious effect on protein A, we sought to determine whether zwitterionic lipids affect the cooperative binding of protein A to RNA1. Thus, we conducted RNA binding experiments under the conditions described in Fig. 3C except that 2 μg/μl MMLs was replaced by 2 μg/μl PC. The RNA binding data was applied to Hill transformation (Fig. 4D) and compared to that in the absence of (Fig. 2G) and presence of MMLs (Fig. 3D). The cooperativity of protein A binding to RNA1 in the presence of PC was similar to that in the absence of MMLs. The presence of PC resulted in little effect on the cooperativity of protein A binding to RNA at low protein concentrations (0.1–2 μM) (Hill coefficient value: 1.342 with PC vs. 1.433 without PC) and high protein concentrations (>2 μM) (Hill coefficient value: 2.7022 with PC vs. 2.429 without MMLs). These results further confirm that protein A-RNA1 interactions are selectively stimulated by specific anionic phospholipids.
MMLs enhance the RNA binding activity of protein A by stimulating its self-interaction
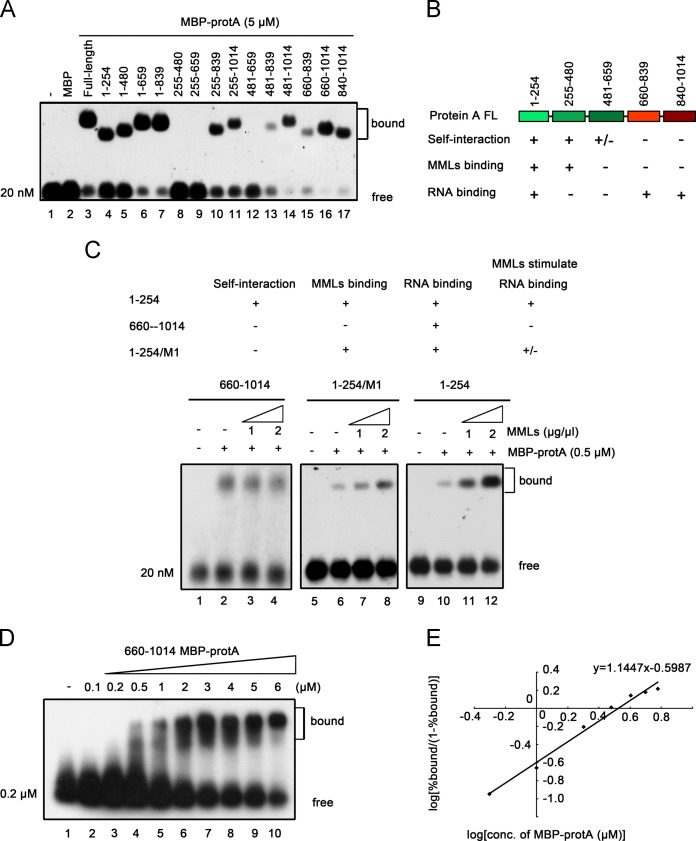
Our previous study showed that protein A is bound to MMLs and protein A self-interaction is enhanced by MMLs (Qiu et al., 2014). Given that RNA binding and self-interaction of protein A are both stimulated by MMLs, we sought to determine the relationship between protein A׳s activities of RNA binding, self-interaction and MML binding. As described previously (Qiu et al., 2014), we constructed and expressed a series of MBP-tagged protein A fragments and then used to determine the fragments responsible for protein A self-interaction and MML binding. We found that protein A fragments amino acids (aa) 1–254 and aa 255–480 are sufficient to mediate protein A self-interaction while aa 481–659 show minor effect on protein A self-interaction (Qiu et al., 2014). Furthermore, aa 1–254 and aa 255–480 are responsible for MML binding in vitro (Qiu et al., 2014).
We mapped the fragments responsible for the RNA binding of protein A by using these protein A fragments in gel mobility shift experiments. Since a major portion of 20 nM RNA probe can be bound by FL protein A at the concentration of 5 μM (Fig. 2B), we used 5 μM each fragment to examine their RNA binding activities at the RNA probe concentration of 20 nM. Our data show that multiple domains, including aa 1–254, aa 660–839, and aa 840–1014, are responsible for the RNA binding of protein A ( Fig. 5A). We then summarized the activities of different protein A fragments. As shown in Fig. 5B, aa 1–254 contains the activities of self-interaction, MML binding and RNA binding. The fragment aa 255–480 contains the activities of MML binding and self-interaction, while aa 660–839 and aa 840–1014 only show the RNA binding activity.
Fig. 5.
MMLs stimulate the RNA binding activity of protein A by promoting its self-interaction. (A) A series of 5 μM MBP-tagged protein A fragments as describe previously (Qiu et al., 2014), were incubated with 20 nM RNA1(50–118) probe and then examined in gel mobility shift assays. (B) Summary the domains of WhNV protein A responsible for self-interaction, MMLs binding and RNA binding, representing the results shown in Fig. 5A and our previous study (Qiu et al., 2014). “+/−” represents that the self-interaction of aa 481–659 is weak (Qiu et al., 2014). (C) The effect of MMLs on the RNA binding of protein A fragments was indicated “+” and “-”, representing the results as indicated below. “+/−” represents that the stimulatory effect of MMLs on the RNA binding is limited. Increasing concentrations of MMLs were incubated with protein A fragments aa 660–1014 (lanes 2–4), 1–254/M1 (lanes 6–8) or 1–254 (lanes 10–12) and RNA1(50–118) probe, and the protein–RNA complex was separated in a gel mobility shift assay. (D,E) Cooperative binding of protein A fragment aa 660–1014 to RNA1(50–118). Gel mobility shift assays were performed using increasing molar concentrations of MBP-protA aa 660–1014 incubated with RNA1(50–118) probe. The Hill coefficients of the RNA binding of protein A at low and high protein concentrations are indicated (E).
Based on this information, it is probable that RNA binding activity of aa 1–254 can be stimulated by MMLs because it contains all the three activities, and the self-interaction activity may be critical for the stimulatory effect of MMLs on the RNA binding activity. To examine this possibility, we incubated the increasing amounts of MMLs with RNA1(50–118) probe and 0.5 μM MBP-protA fragments. As shown in Fig. 5C, increasing MML concentrations dramatically enhanced the RNA binding of protein A aa 1–254 (lanes 9–12), but did not affected the RNA binding of protein A aa 660–1014 (lanes 1–4) due to the lack of membrane-binding activity as expected. Moreover, we used a mutant protein A aa 1–254 fragment, aa 1–254/M1 (K91A, W92A, and R93A), whose MML-binding and RNA-binding activities are intact but self-interaction activity is nulled as described previously (Qiu et al., 2014). Our data show that MMLs only moderately stimulate the RNA binding of aa 1–254/M1 (Fig. 5C, lanes 5–8), indicating that the substantial stimulatory effect of MMLs on the RNA binding requires the self-interaction of protein A. Together, these results indicate that MMLs stimulate RNA binding of protein A via binding to protein A and mediating protein A self-interaction.
To further determine the relationship between the RNA binding and self-interaction of protein A in the absence of MMLs, we examined the cooperativity of protein A fragments aa 660–1014 (Fig. 5D and E). Contrast to the FL protein A, aa 660–1014 has a constant Hill coefficient value of 1.1477, suggesting that the presence of self-interaction enhances the RNA binding of protein A. In summary, we concluded that MMLs stimulate the RNA binding of WhNV protein A by promoting its self-interaction (see Discussion section).
Manipulation of phospholipids metabolism regulates protein A-induced RNA recruitment in cells
To further investigate the effects of MMLs, particularly changes in MMLs, on the RNA binding of protein A in cells, we aimed to manipulate phospholipid synthesis in Pr-E cells to examine protein A׳s activity to recruit RNA1.
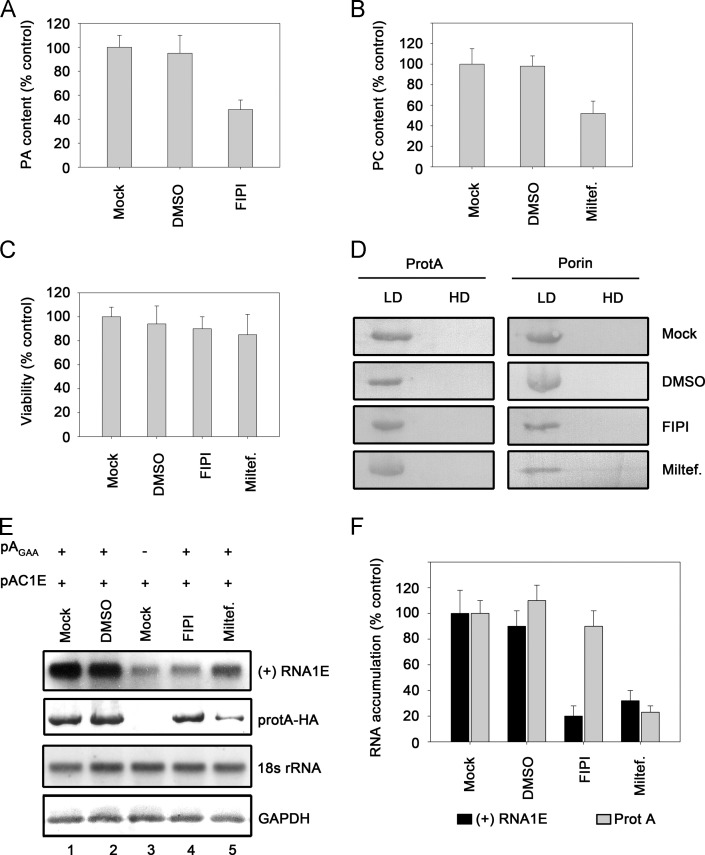
We used PA inhibitor 5-fluoro-2-indolylde-chlorohalopemide (FIPI) and PC inhibitor miltefosine (miltef.) to downregulate PA and PC, respectively, in cells (Castorena et al., 2011, Su et al., 2009). FIPI (100 nM) treatment yielded about 50% reduction in cellular levels of PA ( Fig. 6A), and miltefosine (50 μM) treatment also resulted in about 50% PC reduction (Fig. 6B). Moreover, we also assessed cell viability and found that FIPI and miltefosine moderately affected cell viability (~10% and ~15% reduction, respectively) (Fig. 6C).
Fig. 6.
Manipulation of phospholipids metabolism regulates protein A-induced (+)RNA1E recruitment in cells. (A,B) Measurement of PA and PC content in Pr-E cells treated with 100 nM FIPI (A) and 50 μM miltefosine (B) or with matching concentration of DMSO (vehicle). (C) Viability of cells treated with FIPI, miltefosine or DMSO. (D) FIPI or miltefosine treatment show less effect on the activity of mitochondrial membrane-binding protein to associate with membranes. Nycodenz flotation assay were used to examine membrane association of protein A and porin in cells treated with FIPI, miltefosine or DMSO. LD fractions represent the membrane-rich layers in the gradient, whereas the HD (non-membrane) fractions contain cytosolic soluble proteins. (E) (+)RNA1E accumulation in cells treated with FIPI, miltefosine or DMSO expressing protein AGAA-HA. Cells were divided into two equal fractions. One of fractions was analyzed by Northern blotting with EGFP and 18 s rRNA probes, respectively. The other fraction was analyzed by Western blotting with anti-HA and anti-GAPDH antibodies, respectively. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (F) Quantification data show the accumulation of (+)RNA1E and protein A in Pr-E cells expressing protein AGAA-HA treated with FIPI, miltefosine or DMSO, respectively. The accumulation of RNA and protein is normalized to 18S rRNA and GAPDH, respectively. Error bars represent S.D. values from at least three independent experiments.
Furthermore, we assessed the membrane association of protein A via Nycodenz flotation assay. The flotation gradients were divided into two fractions, LD and HD. As previously described (Qiu et al., 2014), the LD fractions represent the membrane-rich layers in the gradient, whereas the HD (non-membrane) fractions contain cytosolic soluble proteins. WhNV protein A was expressed via transfection with plasmid pAGAA. As shown in Fig. 6D, FIPI or miltefosine treatment did not alter the activity of protein A to associate with membranes. The effect of FIPI or miltefosine treatment on mitochondrial associated protein was also assessed via the detection of porin, which is an integral membrane protein associated with mitochondria. Our results show that FIPI or miltefosine treatment was unable to alter the membrane association of porin as being detected by Nycodenz flotation assay (Fig. 6D), thereby ruling out the possibility that phospholipid inhibitor treatment can damage the property of mitochondrial membranes to associate with membrane-bound proteins.
Next, we examined whether downregulating PA or PC affect protein A׳s activity to recruit RNA1 in cells. Pr-E cells treated with FIPI or miltefosine were transfected with pAGAA and pAC1E. After being transfected for 36 h, Northern blots as well as Western blots were performed to examine the accumulation of (+)RNA1E and protein A, respectively (Fig. 6E). The accumulation of (+)RNA1E in FIPI-treated cells was reduced by about 80% compared to that in mock treated cells (Fig. 6E, “(+)RNA1E”, compared lane 4 to lane 1; Fig. 6F), while protein A accumulation was little affected (Fig. 6E, “protA-HA”, compared lane 3 to lane 1; Fig. 6F), indicating that the changes of the cellular PA levels mediate the RNA binding of protein A in cells. Previous study showed that down-regulating PC synthesis result in the reduction on FHV protein A׳s accumulation through some indirect way (Castorena et al., 2011). Similarly, our findings show that WhNV protein A accumulation was also reduced by 75% in miltefosine-treated cells (Fig. 6E, “protA-HA”, compared lane 5 to lane 1; Fig. 6F). Under the condition that protein A accumulation was substantial reduced, (+)RNA1E accumulation was reduced by 70% as well (Fig. 6E, “(+)RNA1E”, compared lane 5 to lane 1; Fig. 6F).
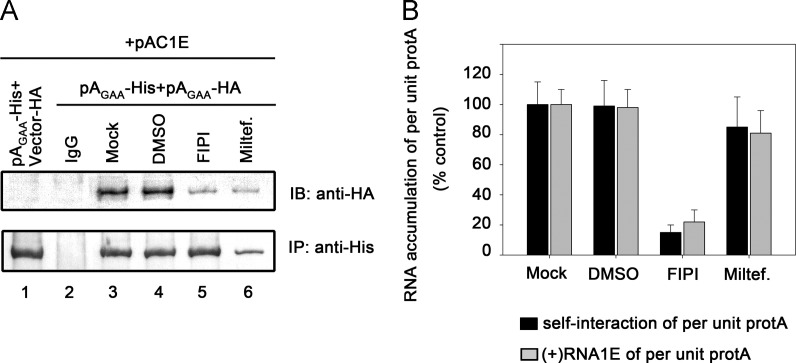
To exclude the effects of protein A accumulation on (+)RNA1E accumulation, we sought to determine the activity of per unit protein A to recruit (+)RNA1E. Because the RNA binding and self-interaction of protein A were closely linked, the self-interaction of protein A in cells was also examined. To this end, Protein AGAA with His or HA tags was expressed in the presence of (+)RNA1E. Protein AGAA-His expressing in different phospholipid inhibitors-treated cells was immunoprecipitated and the accumulation of (+)RNA1E recruited by the immunoprecipitated protein AGAA was analyzed by real-time RT-PCR. The relationship between protein A recruiting (+)RNA1E and self-interaction was also determined by examining the coimmunoprecipitated Protein AGAA-HA. As shown in Fig. 7A, miltefosine treatment resulted in substantial reduction on protein A accumulation (“IP”, compared lane 6 to lane 3), while FIPI treatment show limited effect on protein A accumulation (“IP”, compared lane 5 to lane 3). The self-interaction of protein A was also substantially reduced in both FIPI and miltefosine-treated cells (Fig. 7A, “IB”, compared lanes 5 or 6 to lane 3).
Fig. 7.
The activities of per unit protein A recruiting (+)RNA1E and self-interaction are regulated by manipulation of phospholipids in cells. (A) Pr-E cells expressing (+)RNA1E together with protein AGAA-His plus empty vector-HA (lane 1) or protein AGAA-His plus protein AGAA-HA (lanes 2–6) were harvested. Cell Lysates were immunoprecipitated with an anti-His antibody (lanes 1 and 3–6) or control IgG (lane 2). The immunoprecipitated complexes were divided into two equal fractions. One of fractions was blotted with an anti-HA antibody to determine the self-interaction of protein A. The (+)RNA1E associated with the immunoprecipitated protein A was separated from the other fraction and following analyzed by real-time RT-PCR. (B) Graph of the ratios of the accumulation of (+)RNA1E analyzed by real-time RT-PCR as described above and protein A self-interaction versus protein A׳s accumulation. The accumulation of RNA and protein is normalized to 18S rRNA and GAPDH from total cells, respectively. Error bars represent S.D. values from at least three independent experiments.
We determined the activities of per unit protein A by calculating the ratio of the levels of (+)RNA1E recruited by the immunoprecipitated protein A and protein A self-interaction to that of protein A (Fig. 7B). As the PA concentration decreased, the activities of per unit protein A to recruit (+)RNA1E and self-interact were correspondingly reduced by 78% and 82%, respectively (Fig. 7B, “FIPI”). These results were consistent with the in vitro data that PA mediate RNA binding of protein A (Fig. 4) as well as our previous study that manipulation of PA biosynthesis mediates protein A self-interaction in cells (Qiu et al., 2014).
In contrast, as the PC concentration decreased, the activity of per unit protein A to recruit (+)RNA1E was only reduced by 19% (Fig. 7B, “Miltef.”) and the activity of per unit protein A to self-interact was reduced by 15% (Fig. 7B, “Miltef.”). These results indicate that the reduction in cellular PC levels show limited effects on per unit protein A activities, which were consistent with the in vitro data that PC showed limited effect on protein A binding to RNA (Fig. 4) as well as our previous study that PC show limited effect on protein self-interaction (Qiu et al., 2014). The observations that the activities of per unit protein A to recruit (+)RNA1E and self-interact were tightly linked in different phospholipid inhibitors-treated cells further confirm the relationship between protein A binding to RNA and self-interaction. Altogether, from this set of experiments, we concluded that different phospholipids specifically regulate the activity of protein A (per unit of protein A) recruiting genomic RNA1, and this regulation is accomplished by MMLs mediating protein A self-interaction.
Discussion
RNA replication of (+)RNA viruses requires the association of viral RNA and replicases with intracellular membranes to form vRCs (Ahlquist, 2006, Ahlquist et al., 2003, Miller and Krijnse-Locker, 2008, Sasvari and Nagy, 2010). To advance the understanding of the relationship between intracellular membranes and viral RNA replicases, we studied the direct effects of MMLs on the RNA binding of WhNV protein A. We uncover that WhNV protein A binds to RNA1 in a cooperative manner, and the cooperativity could be stimulated by high protein A concentrations. MMLs stimulated the RNA-binding activity of protein A, and this stimulation exhibited strong selectivity for different phospholipids. Interestingly, MMLs stimulated the RNA-binding cooperativity only at higher protein A concentrations. Furthermore, manipulating phospholipid metabolisms significantly inhibited protein A-induced RNA1 recruitment/stabilization in cells. Altogether, these findings demonstrate the important role of MMLs in the RNA binding activity of WhNV protein A to properly function as a viral RNA replicase.
For (+)RNA viruses, recruiting RNA to membranes by viral replicases is an essential step prior to RNA replication (Pogany et al., 2005, Schwartz et al., 2002, Sullivan and Ahlquist, 1999, Van Wynsberghe and Ahlquist, 2009, Van Wynsberghe et al., 2007). The direct RNA–protein interaction was stimulated by MMLs (Fig. 3A) indicating that MMLs play a direct role in regulating the activity of WhNV protein A to recruit viral genomic RNA. Binding to RNA in a cooperative manner can be advantageous for genomic RNA recruitment, since this may increase the stability of protein–RNA complexes in cells. Therefore, many viral proteins, including NS5B of HCV (Wang et al., 2002), 3D of poliovirus (Arnold and Cameron, 1999), p33 of Tomato bushy stunt virus (Rajendran and Nagy, 2003) and N protein of severe acute respiratory syndrome coronavirus (Chang et al., 2009) were all found to bind viral RNAs cooperatively. The novel findings of this current work are that the cooperativity of WhNV protein A binding to RNA1 can be stimulated by MMLs and, very interestingly, this stimulation takes effect only at high protein A concentrations (Figs. 2G and 3D; Hill co-efficient: 6.613 vs 2.429 at high concentrations; 1.477 vs 1.433 at low concentrations). The functional significance of this phenomenon is currently not known. A reasonable speculation is that MMLs regulate viral replication by modulating the distribution of viral genomic RNA between translational machinery and vRCs during different stages of viral infection. At a very early stage, few viral genomic RNAs and replicases exist, and the priority of nodaviral RNA1 is to function as mRNA to translate sufficient protein A. As more and more protein A molecular is translated from RNA1 and moves to membranes, resulting in the increased density of protein A on membranes, the priority of RNA1 subsequently shifts to replicate more viral RNAs. Thus, at this stage, the cooperative binding of protein A to RNA1 was substantially stimulated by MMLs, thereby resulting in the recruitment of otherwise cytoplasmic RNA1 to membranes to form vRCs.
Most (+)RNA viruses typically encode multiple viral replicase proteins, and their intermolecular interactions are critical for RNA replication (Diaz et al., 2012, Dye et al., 2005, Panavas et al., 2005, Panaviene et al., 2003, Wang et al., 2002). For nodavirus, the self-interaction of protein A, the sole replicase, is required for RNA replication (Dye et al., 2005). Moreover, three-dimensional analysis showed that FHV protein A forms higher-order oligomers in spherules (Kopek et al., 2007). These results indicated that the polymerization of nodaviral protein A is required for RNA replication. However, the precise role of polymerization/self-interaction is currently not known. The study reported here highlighted the relationship between the RNA binding and self-interaction of nodaviral protein A. Multiple lines of evidence indicate that MMLs stimulate the RNA binding of protein A to RNA by promoting its self-interaction. First, we showed that protein A aa 1–254/M1 mediates RNA binding and MML binding, whereas protein A aa 1–254 are responsible for RNA binding, MML binding and protein A self-interaction (Fig. 5C). The RNA binding of protein A aa 1–254/M1 was moderately affected by MMLs, whereas the RNA binding of protein A aa 1–254 was substantially stimulated by MMLs (Fig. 5C), indicating that the substantial stimulatory effect of MMLs on the RNA binding requires protein A self-interaction. However, the contribution of the direct MML-protein A interaction to the RNA binding cannot be omitted but may not be the main reason for the substantial stimulation of MMLs on protein A binding to RNA. Furthermore, we found that higher concentrations of FL protein A stimulated the cooperativity of its binding to RNA even in the absence of MMLs (Fig. 2G). Given that protein–RNA interaction occurred in a pure reaction solution, which contains only RNA probe and protein A, the higher concentration-induced increase in protein A self-interaction should be the cause of the enhanced cooperativity of protein A binding to RNA1. Therefore, our findings indicate that MMLs stimulate protein A self-interaction, which in turn substantially promotes the efficiency of the cooperative binding of protein A to RNA. The activities of per unit protein A recruiting (+)RNA1E and self-interaction are tightly linked in cells further confirm the relationship between these two activities of protein A (Fig. 7).
Membrane lipids comprise certain phospholipids, and the composition of these phospholipids is different for different membranes (van Meer et al., 2008). Many previous studies showed that certain phospholipids have different effects on different (+)RNA replicases. The particular phospholipids enriched in certain intercellular membranes, which are associated with different viruses, show preferential and direct effects on the activities of replicases. According to nodavirus, WhNV protein A activities (Qiu et al., 2014) (Fig. 4) can be mediated by specific anionic phospholipids CL, PA and PG that are enriched in mitochondrial membranes (van Meer et al., 2008), the sites of which WhNV and FHV protein As are associated with (Miller and Ahlquist, 2002, Miller et al., 2001, Qiu et al., 2013). These certain lipids may mediate protein A activities by partitioning protein A into liposome fraction and thus leading to the increase of protein A׳s local density. Indeed, the RNA binding to protein A is cooperative, and the cooperativity could be enhanced by higher protein A concentration (Fig. 2G), indicating that protein As show stronger RNA binding activity when they are gathered together.
In contrast to the in vitro data that MMLs showed obvious effects on WhNV protein A binding to RNA (Fig. 3, Fig. 4, Fig. 5), down-regulating the phospholipid synthesis in cells result in relatively minor effects on protein A activity (Fig. 6, Fig. 7). That may be partly due to the fact that the simplified in vitro systems cannot represent the complexity of the protein A in cells. In addition, the possible interference of MBP tag to the activity of protein A still remains. Future studies will focus on solving these limitations of the methods and determining the exact and complete activity of protein A.
Overall, our results show that MMLs directly regulate the activity of WhNV protein A binding to RNA1. This regulation requires the activities of protein A binding to MMLs and self-interaction, and could be specifically moderated by certain phospholipids. These results highlight the detailed mechanisms by which intracellular membranes regulate the functions of nodaviral replicase protein A and provide new insights for further study of the role of intracellular membranes in the RNA replication of (+)RNA viruses.
Materials and methods
Plasmids
Standard procedures were used for restriction of nuclease digestion and plasmid DNA construction and purification. To analyze WhNV protein A activity in cells, protein A ORF and RNA1 was inserted into pAC5.1/V5-His B vector (Invitrogen, Carlsbad, CA). Plasmids for the purification of MBP fusion protein A were constructed by inserting protein A ORF into pMAL-c2X (New England BioLabs, Ipswich, MA). Mutations were introduced into protein A ORF via PCR-mediated mutagenesis as described previously (Qiu et al., 2011, Qiu et al., 2014, Qiu et al., 2013).
Cells and transfection
Pr-E cells which are derived from P. rapae larvae, the natural host of WhNV, and were successfully utilized to study WhNV RNA replication previously (Qiu et al., 2011, Qiu et al., 2014, Qiu et al., 2013), were maintained at 27 °C in Grace׳s medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Gibco). DNA plasmids were transfected into cells using FuGENE HD transfection reagent (Roche, Basel, Switzerland) according to the manufacturer׳s protocol. All subsequent assays were performed 36 h after transfection except where indicated otherwise.
Western blot analysis and antibodies
The proteins extracted from cells were subjected to 10% SDS-PAGE and Western blot analysis as previously described (Qiu et al., 2011, Qiu et al., 2014, Qiu et al., 2013). Unless otherwise indicated, the anti-MBP polyclonal antibody was purchased from New England BioLabs, and the other primary and secondary antibodies were purchased from Proteintech, Chicago, IL, USA.
RNA extraction, northern blot analysis, and TaqMan real-time RT-PCR
Total RNA was extracted from cells or cell lysates using TRIzol reagent (Invitrogen) and digested with RQ1 RNase-free DNase I (Promega, Madison, WI, USA) as previously described (Qiu et al., 2011, Qiu et al., 2014, Qiu et al., 2013). For Northern blot analysis, 2 μg of each RNA sample was analyzed via Northern blot analysis as previously described (Qiu et al., 2011, Qiu et al., 2014, Qiu et al., 2013). The probes for (+)EGFP were complementary to the entire EGFP sequences. All probes were labeled with DIG-UTP (Roche) for in vitro transcription. For real-time PCR, 5 μg of RNA was subjected to reverse transcription with Superscript III reverse transcriptase (Invitrogen) using random hexamer primers (Invitrogen). The total cDNAs were then used for real-time PCR using TaqMan probes targeted for EGFP and 18S rRNA, respectively (Qiu et al., 2013). PCR was carried out at 98 °C for 3 min, followed by 45 cycles at 98 °C for 15 s, 57 °C for 10 s, and 68 °C for 10 s. The identical temperature profiles were used for all real-time RT-PCR runs and fluorescence values were collected during the annealing step. All samples were analyzed in triplicate and were evaluated in at least three independent experiments.
Purification of protein A and its derivatives
The expression and purification of recombinant WhNV protein A and its derivatives were carried out as previously described (Qiu et al., 2014, Wu et al., 2014). Briefly, to obtain soluble recombinant protein, MBP-tagged FL protein A and its mutants as well as the negative control protein MBP were expressed in E. coli strain TB1 at 20 °C in the presence of 0.2 mM IPTG. Cell pellets were resuspended in binding buffer (20 mM Tris–HCl [pH 7.4], 200 mM NaCl, 1 mM EDTA, 10 mM 2-Mercaptoethanol) supplemented with 1.5% Triton-X 100 and protease inhibitors cocktail (Sigma, St. Louis, Mo, USA). Cells were lysed by sonication and then debris was removed by centrifugation for 30 min at 11,000g. The proteins in the supernatant were purified using amylose resin (New England BioLabs) according to the manufacturer׳s protocol and concentrated using Amicon Ultra-15 filters (Millipore, Schwalbach, Germany), and the buffer was exchanged to the hypotonic buffer (1 mM HEPES [pH 7.4], 0.1 mM EDTA, 15 mM NaCl, 1 mM DTT).
Mitochondrial membrane lipids and liposomes
Mitochondrial outer membranes were isolated from Pr-E cells by mechanical disruption and differential centrifugation as previously described (Graham, 1993, Graham, 2001). Subsequently, the purified outer mitochondrial membranes were treated with 0.1 mg/ml proteinase K (Sigma) for 10 min in hypotonic buffer supplemented with 1.5% Triton-X 100 to dissolve integral membrane proteins. MMLs were then reisolated by centrifugation at 12,000g for 20 min and resuspended in hypotonic buffer. MMLs were further purified and concentrated by using Amicon Ultra-15 filters (Millipor). Lipids were obtained from Sigma in the highest purity grades available: CL, PA, PG, PS, PC and PE. The liposomes were prepared as described (Ahola et al., 1999, Stapleford et al., 2009, Wu et al., 1992, Wu and Kaesberg, 1991). Briefly, the purchased lipids were dissolved and mixed in chloroform/methanol (2:1) at 10 mg lipid per 1 ml organic solvent. The mixture was dried under nitrogen and lyophilized to remove any traces of solvent. The dry film was hydrated with 20 mM HEPES buffer at pH 7.4 by vortexing overnight at 4 °C. The purified MMLs and liposomes were quantified by Bradford protein assay (Bio-Rad) using a UV–visible spectrophotometer (Shimadzu).
Membrane flotation assays
Cells were recovered via scraping and centrifugation, resuspended in hypotonic buffer, and spun at 4 °C for 15 min. Unbroken cells, nuclei, and large debris were removed via centrifugation at 500g for 5 min to obtain the initial total lysates. Nycodenz (Sigma) was added to the total lysates to a final concentration of 37.5% (wt/vol), and samples were loaded under a 5–25% discontinuous Nycondenz gradient prepared in hypotonic buffer and centrifuged to equilibrium at 100,000g for 20 h at 4 °C in a Beckman coulter SW40 rotor. After centrifugation, the gradient was divided into the upper half of the gradient (LD fraction) and the lower half of the gradient (HD fraction). Protein samples were isolated from half of each fraction via centrifugation at 180,000g in a Beckman coulter SW40 rotor for 3 h and then analyzed via Western blotting.
Gel mobility shift assay
Briefly, MBP-tagged protein A was incubated with 20 nM DIG-labeled RNA1(50–118) probe in a binding buffer (50 mM HEPES [pH 7.4], 15 mM NaCl, 10% glycerol, and 1.5 U of RNase inhibitor [promega]) at 27 °C for 30 min; the total volume was 10 μl. To test the stimulating effects of MMLs on protein A binding to RNA, the RNA probe was increased to 0.2 μM, and MMLs were added (0.1–10 μg MMLs per 1 μl reaction mixture) in the reaction mixtures with the MBP-tagged protein A. For competition experiments, unlabeled competitors (applied in molar excess as indicated in Fig. 2) were added simultaneously with the labeled RNA probe to the binding reaction. After the binding reaction, the samples were analyzed via 1.0% or 2.0% agarose gel electrophoresis run at 100 V in 0.5× TBE buffer in a cold room and transferred to Hybond N nylon membranes via capillary transfer. The membranes were incubated for 30 min with anti-DIG antibody conjugated with alkaline phosphatase (Roche), exposed to film. The RNA probe representing the RNA1(50–118) was labeled with DIG-UTP for in vitro transcription. The values of Hill coefficient, an indicator of the cooperativity of RNA-binding, were calculated using Hill transformation of the RNA-binding data obtained in repeated experiments. An apparent dissociation constant (Kd) for protein–RNA interaction, the concentration of the protein at which 50% of RNA was retarded, was also determined on the basis of these data.
Coimmunoprecipitation assays
Coimmunoprecipitation assays were performed as previously described (Qi et al., 2011, Qi et al., 2012, Qiu et al., 2013). Briefly, 36 h after transfection, cells were lysed with NETN buffer [20 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, and 0.5% NP-40] for 20 min at 4 °C in the presence of protease inhibitors cocktail (Sigma). Lysates were clarified at 12,000 rpm for 10 min at 4 °C, and then postnuclear lysates were precleared via incubation with protein-G agarose beads (Roche) coupled to goat anti-mouse IgG and then incubated with mouse anti-His antibody at room temperature for 4 h. The antibody-bound complexes were captured, washed, and divided into two equal fractions. One of the fractions was then subjected to SDS-PAGE and Western blotting analysis with rabbit anti-His antibody. The RNAs associated with the immunoprecipitated complexes were separated from the other fraction and following analyzed by real-time RT-PCR as described above.
PA inhibitor and cells viability assays
FIPI (Sigma) and miltefosine (Sigma) was used to inhibit PA and PC production as previously described (Castorena et al., 2011, Su et al., 2009). Briefly, 12 h after transfection, cells were treated with 75 nM FIPI or 50 μM in DMSO and incubated for another 24 h. Total PA and PC content was determined using the modified phospholipase D-based enzymatic method as previously described (Hojjati and Jiang, 2006, Qiu et al., 2014). Cell viability assays were performed using MTT (Sigma) as previously described (Castorena et al., 2011).
Acknowledgments
We are grateful to Profs. Zehua Yu and Just Vlak for generously providing experimental materials. We also thank Dr. Jiali Si for critically reading and Ms. Markeda Wade for professionally editing this manuscript.
This work was supported by the National Basic Research Program of China (973 Program, 2014CB542603), the National Natural Science Foundation of China grants No. 31270190 (to X.Z.), No. 81201292 (to X.Z.) and No. 31270189 (to Y.H.), and the Chinese 111 Project Grant no. B06018.
References
- Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 2006;4:371–382. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist P., Noueiry A.O., Lee W.M., Kushner D.B., Dye B.T. Host factors in positive-strand RNA virus genome replication. J. Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahola T., Lampio A., Auvinen P., Kaariainen L. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. Embo. J. 1999;18:3164–3172. doi: 10.1093/emboj/18.11.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J.J., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro. J. Biol. Chem. 1999;274:2706–2716. doi: 10.1074/jbc.274.5.2706. [DOI] [PubMed] [Google Scholar]
- Ball L.A. Requirements for the self-directed replication of flock house virus RNA 1. J. Virol. 1995;69:720–727. doi: 10.1128/jvi.69.2.720-727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castorena K.M., Stapleford K.A., Miller D.J. Complementary transcriptomic, lipidomic, and targeted functional genetic analyses in cultured Drosophila cells highlight the role of glycerophospholipid metabolism in Flock House virus RNA replication. BMC Genomics. 2011;11:183. doi: 10.1186/1471-2164-11-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Hsu Y.L., Chang Y.H., Chao F.A., Wu M.C., Huang Y.S., Hu C.K., Huang T.H. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol. 2009;83:2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V., Heaton N.S., Randall G. Lipids at the interface of virus–host interactions. Curr. Opin. Microbiol. 2012;15:512–518. doi: 10.1016/j.mib.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A., Gallei A., Ahlquist P. Bromovirus RNA replication compartment formation requires concerted action of 1a׳s self-interacting RNA capping and helicase domains. J. Virol. 2012;86:821–834. doi: 10.1128/JVI.05684-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye B.T., Miller D.J., Ahlquist P. in vivo self-interaction of nodavirus RNA replicase protein a revealed by fluorescence resonance energy transfer. J. Virol. 2005;79:8909–8919. doi: 10.1128/JVI.79.14.8909-8919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froshauer S., Kartenbeck J., Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T.M., Friesen P.D., Rueckert R.R. Autonomous replication and expression of RNA 1 from black beetle virus. J. Virol. 1983;46:481–489. doi: 10.1128/jvi.46.2.481-489.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J.M. Isolation of mitochondria, mitochondrial membranes, lysosomes, peroxisomes, and Golgi membranes from rat liver. Methods Mol. Biol. 1993;19:29–40. doi: 10.1385/0-89603-236-1:29. [DOI] [PubMed] [Google Scholar]
- Graham J.M. Isolation of mitochondria from tissues and cells by differential centrifugation. Curr Protoc Cell Biol. 2001;Chapter 3(Unit 3):3. doi: 10.1002/0471143030.cb0303s04. [DOI] [PubMed] [Google Scholar]
- Heaton N.S., Randall G. Multifaceted roles for lipids in viral infection. Trends Microbiol. 2011;19:368–375. doi: 10.1016/j.tim.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojjati M.R., Jiang X.C. Rapid, specific, and sensitive measurements of plasma sphingomyelin and phosphatidylcholine. J. Lipid Res. 2006;47:673–676. doi: 10.1194/jlr.D500040-JLR200. [DOI] [PubMed] [Google Scholar]
- Jonczyk M., Pathak K.B., Sharma M., Nagy P.D. Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology. 2007;362:320–330. doi: 10.1016/j.virol.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Kampmueller K.M., Miller D.J. The cellular chaperone heat shock protein 90 facilitates Flock House virus RNA replication in Drosophila cells. J. Virol. 2005;79:6827–6837. doi: 10.1128/JVI.79.11.6827-6837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopek B.G., Perkins G., Miller D.J., Ellisman M.H., Ahlquist P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 2007;5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujala P., Ikaheimonen A., Ehsani N., Vihinen H., Auvinen P., Kaariainen L. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 2001;75:3873–3884. doi: 10.1128/JVI.75.8.3873-3884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.M., Ishikawa M., Ahlquist P. Mutation of host delta9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J. Virol. 2001;75:2097–2106. doi: 10.1128/JVI.75.5.2097-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Li W.X., Ding S.W. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- Magliano D., Marshall J.A., Bowden D.S., Vardaxis N., Meanger J., Lee J.Y. Rubella virus replication complexes are virus-modified lysosomes. Virology. 1998;240:57–63. doi: 10.1006/viro.1997.8906. [DOI] [PubMed] [Google Scholar]
- Mas P., Beachy R.N. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J. Cell Biol. 1999;147:945–958. doi: 10.1083/jcb.147.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.J., Ahlquist P. Flock house virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J. Virol. 2002;76:9856–9867. doi: 10.1128/JVI.76.19.9856-9867.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.J., Schwartz M.D., Ahlquist P. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 2001;75:11664–11676. doi: 10.1128/JVI.75.23.11664-11676.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S., Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohturfft A., Zhang S.C. Coordination of lipid metabolism in membrane biogenesis. Annu. Rev. Cell Dev. Biol. 2009;25:539–566. doi: 10.1146/annurev.cellbio.24.110707.175344. [DOI] [PubMed] [Google Scholar]
- Panavas T., Hawkins C.M., Panaviene Z., Nagy P.D. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology. 2005;338:81–95. doi: 10.1016/j.virol.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Panaviene Z., Baker J.M., Nagy P.D. The overlapping RNA-binding domains of p33 and p92 replicase proteins are essential for tombusvirus replication. Virology. 2003;308:191–205. doi: 10.1016/s0042-6822(02)00132-0. [DOI] [PubMed] [Google Scholar]
- Pathak K.B., Pogany J., Nagy P.D. Non-template functions of the viral RNA in plant RNA virus replication. Curr. Opin. Virol. 2011;1:332–338. doi: 10.1016/j.coviro.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Pathak K.B., Pogany J., Xu K., White K.A., Nagy P.D. Defining the roles of cis-acting RNA elements in tombusvirus replicase assembly in vitro. J. Virol. 2012;86:156–171. doi: 10.1128/JVI.00404-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K.W., van der Meer Y., Roos N., Snijder E.J. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 1999;73:2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J., White K.A., Nagy P.D. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 2005;79:4859–4869. doi: 10.1128/JVI.79.8.4859-4869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi N., Cai D., Qiu Y., Xie J., Wang Z., Si J., Zhang J., Zhou X., Hu Y. RNA binding by a novel helical fold of b2 protein from wuhan nodavirus mediates the suppression of RNA interference and promotes b2 dimerization. J. Virol. 2011;85:9543–9554. doi: 10.1128/JVI.00785-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi N., Zhang L., Qiu Y., Wang Z., Si J., Liu Y., Xiang X., Xie J., Qin C.F., Zhou X., Hu Y. Targeting of dicer-2 and RNA by a viral RNA silencing suppressor in Drosophila cells. J. Virol. 2012;86:5763–5773. doi: 10.1128/JVI.07229-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Cai D., Qi N., Wang Z., Zhou X., Zhang J., Hu Y. Internal initiation is responsible for synthesis of Wuhan nodavirus subgenomic RNA. J. Virol. 2011;85:4440–4451. doi: 10.1128/JVI.02410-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Wang Z., Liu Y., Han Y., Miao M., Qi N., Yang J., Xia H., Li X.F., Qin C.F., Hu Y., Zhou X. The self-interaction of a Nodavirus replicase is enhanced by mitochondrial membrane lipids. PLoS One. 2014;9:e89628. doi: 10.1371/journal.pone.0089628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Wang Z., Liu Y., Qi N., Miao M., Si J., Xiang X., Cai D., Hu Y., Zhou X. Membrane association of Wuhan nodavirus protein A is required for its ability to accumulate genomic RNA1 template. Virology. 2013;439:140–151. doi: 10.1016/j.virol.2013.02.010. [DOI] [PubMed] [Google Scholar]
- Rajendran K.S., Nagy P.D. Characterization of the RNA-binding domains in the replicase proteins of tomato bushy stunt virus. J. Virol. 2003;77:9244–9258. doi: 10.1128/JVI.77.17.9244-9258.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasvari Z., Nagy P.D. Making of viral replication organelles by remodeling interior membranes. Viruses. 2010;2:2436–2442. doi: 10.3390/v2112436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel A., Giddings T.H., Jr., Ladinsky M.S., Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Mende J., Bieck E., Hugle T., Penin F., Rice C.M., Blum H.E., Moradpour D. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 2001;276:44052–44063. doi: 10.1074/jbc.M103358200. [DOI] [PubMed] [Google Scholar]
- Schneemann A., Zhong W., Gallagher T.M., Rueckert R.R. Maturation cleavage required for infectivity of a nodavirus. J. Virol. 1992;66:6728–6734. doi: 10.1128/jvi.66.11.6728-6734.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M., Chen J., Janda M., Sullivan M., den Boon J., Ahlquist P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell. 2002;9:505–514. doi: 10.1016/s1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- Stapleford K.A., Rapaport D., Miller D.J. Mitochondrion-enriched anionic phospholipids facilitate flock house virus RNA polymerase membrane association. J. Virol. 2009;83:4498–4507. doi: 10.1128/JVI.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Yeku O., Olepu S., Genna A., Park J.S., Ren H., Du G., Gelb M.H., Morris A.J., Frohman M.A. 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol. Pharmacol. 2009;75:437–446. doi: 10.1124/mol.108.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M.L., Ahlquist P. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 1999;73:2622–2632. doi: 10.1128/jvi.73.4.2622-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe P.M., Ahlquist P. 5′ cis elements direct nodavirus RNA1 recruitment to mitochondrial sites of replication complex formation. J. Virol. 2009;83:2976–2988. doi: 10.1128/JVI.02040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe P.M., Chen H.R., Ahlquist P. Nodavirus RNA replication protein a induces membrane association of genomic RNA. J. Virol. 2007;81:4633–4644. doi: 10.1128/JVI.02267-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter P.A., Schneemann A. Recent insights into the biology and biomedical applications of Flock House virus. Cell Mol. Life Sci. 2008;65:2675–2687. doi: 10.1007/s00018-008-8037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.M., Hockman M.A., Staschke K., Johnson R.B., Case K.A., Lu J., Parsons S., Zhang F., Rathnachalam R., Kirkegaard K., Colacino J.M. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 2002;76:3865–3872. doi: 10.1128/JVI.76.8.3865-3872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Qiu Y., Liu Y., Qi N., Si J., Xia X., Wu D., Hu Y., Zhou X. Characterization of a nodavirus replicase revealed a de novo initiation mechanism of RNA synthesis and terminal nucleotidyltransferase activity. J. Biol. Chem. 2013;288:30785–30801. doi: 10.1074/jbc.M113.492728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L., Hirata Y., Arai M., Kohara M., Wakita T., Watashi K., Shimotohno K., He Y., Zhong J., Toyoda T. Sphingomyelin activates hepatitis C virus RNA polymerase in a genotype-specific manner. J. Virol. 2010;84:11761–11770. doi: 10.1128/JVI.00638-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K.A., Nagy P.D. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:187–226. doi: 10.1016/S0079-6603(04)78005-8. [DOI] [PubMed] [Google Scholar]
- Wu S.X., Ahlquist P., Kaesberg P. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc. Natl. Acad. Sci. USA. 1992;89:11136–11140. doi: 10.1073/pnas.89.23.11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.X., Kaesberg P. Synthesis of template-sense, single-strand Flockhouse virus RNA in a cell-free replication system. Virology. 1991;183:392–396. doi: 10.1016/0042-6822(91)90153-3. [DOI] [PubMed] [Google Scholar]
- Wu W., Wang Z., Xia H., Liu Y., Qiu Y., Hu Y., Zhou X. Flock house virus RNA polymerase initiates RNA synthesis de novo and possesses a terminal nucleotidyl transferase activity. PLoS One. 2014;9:e86876. doi: 10.1371/journal.pone.0086876. [DOI] [PMC free article] [PubMed] [Google Scholar]