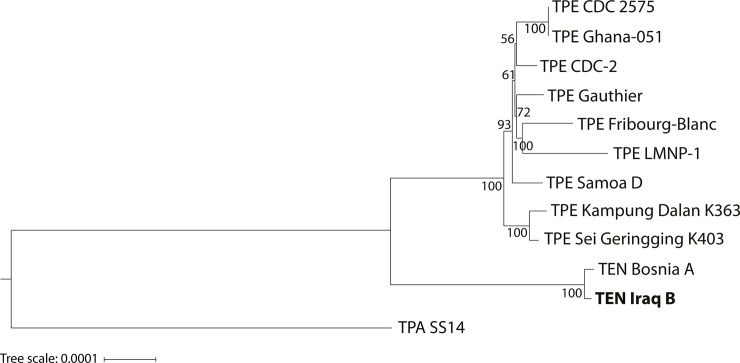
Fig 1. A phylogenetic tree based on the alignment of the Treponema pallidum subsp. endemicum (TEN) Iraq B genome with additional TEN and T. pallidum subsp. pertenue (TPE) genomes.
The tree was constructed from the complete genome sequences of the TEN strains (Bosnia A, Iraq B) and TPE strains (CDC-2, Gauthier, Samoa D, Ghana-051, CDC 2575, Kampung Dalan K363, Sei Geringging K403, LMNP-1, and Fribourg-Blanc). The tprK sequences, tRNA-Ile and tRNA-Ala regions of both rRNA operons, tprD, arp, and TP0470 genes were omitted from the analysis due to their recombinant or repetitive character. The genome sequence of the T. pallidum subsp. pallidum (TPA) strain SS14 was used as an outgroup. There were a total of 1,129,405 positions in the final dataset. The Maximum Likelihood tree was constructed in RAxML-NG (v0.9.0) using the TN93 substitution model [23]. Gamma-distributed substitution rates among sites and proportion of invariable sites were applied. The robustness of the tree branches was assessed by 500-bootstrap replicate analyses. Predicted tree was visualized by iTOL (v5.5) [25]. The bar scale corresponds to a difference of 0.0001 nucleotides per nucleotide site.