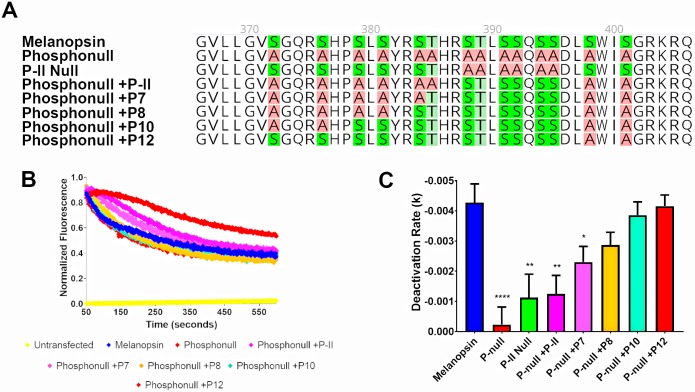
Fig 3. At least eight proximal C-terminus phosphorylation sites are needed to deactivate melanopsin.
(A) 2D schematic of melanopsin and phosphonull mutants analyzed in this figure. (B) Representative calcium imaging of transient melanopsin-expressing HEK293 cells expressing melanopsin and phosphonull mutants with varying numbers of proximal C-terminus phosphorylation sites mutated back to serines or threonines. (C) Quantified deactivation rates of all melanopsin mutants examined in this experiment. Error bars represent S.E.M. of three independent transfections. All mutants’ rates were compared to wild-type melanopsin’s rate. P<0.05, 0.01, 0.001, 0.0001 correspond to *, **, ***, ****, respectively. 2D schematic of melanopsin sequences made using Geneious software [53].