Figure 3.
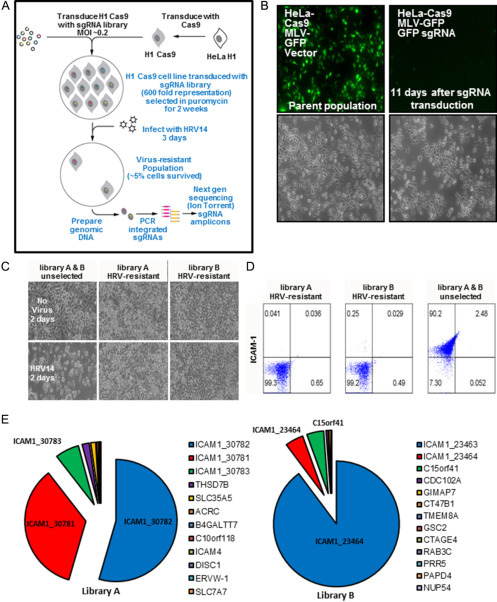
CRISPR/Cas9 screen for HRV host factors. (A) The HRV-HF CRISPR/Cas9 screen workflow showing the generation of the Cas9 expressing H1-HeLa cells containing the sgRNA libraries followed by their subsequent challenge with HRV14 and the assessment of the enriched sgRNAs using next-gen sequencing. (B) HeLa-H1-Cas9 cells were transduced with Moloney Leukemia virus (MLV)-GFP, then supra-transduced with either an empty vector control (parent population) or one expressing a sgRNA against GFP. The cells were selected for puromycin resistance and cultured for 11 days then fixed and imaged for GFP expression. Differential interference contrast (DIC) images are provided below. 4 × magnification. (C) DIC images of cells transduced with either library A or B that survived the HRV14 challenge were expanded and tested for their susceptibility to HRV14’s cytopathic effect over 2 days (bottom row) compared to the unselected parent cell population and the respective uninfected cell populations (top row). (D) Cells from (C) were fixed and immunostained for ICAM1 surface expression by flow cytometry. (E) A chart showing the relative proportion of total sequencing reads for the recovered sgRNAs from the HRV14 CRISPR/Cas9 pooled screen based upon the analysis of genomic DNA from the surviving cells from library A or B. Gene names are provided for each sgRNA with the associated numbers designating their unique identifying library number.
