Fig. 1.
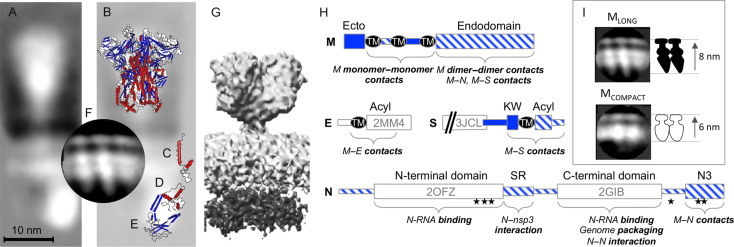
Structure and organization of proteins in the virion. Cryo-EM reconstruction of the SARS-CoV structural proteins from virions (A; Neuman et al., 2006) superimposed with solved structures of the MHV S ectodomain (B; 3JCL), SARS-CoV E (C; 2MM4), SARS-CoV C-terminal domain (D; 2GIB), SARS-CoV N-terminal domain (E; 2OFZ), and a cryo-EM reconstruction of the M proteins from MHV VLPs (F; Neuman et al., 2011). An alternative shorter, wider cryo-EM reconstruction of SARS-CoV virion proteins is shown for comparison (G; EMD-1423). Schematics are based on MHV proteins, following the annotation of Kuo et al. (2016). The orientation of N protein domains shown here is hypothetical, and is intended for illustrative purposes only. (H) Domain structure and annotation of MHV S, E, M, and N proteins showing domains outside the virion (solid blue), inside the virion (striped blue), and the position of solved structures. Transmembrane regions (TM), sites of palmitoylation (Acyl), a conserved sequence preceding the transmembrane of S (KW), a serine–arginine-rich unstructured region (SR), phosphorylation sites (stars), and the C-terminal M-interacting domain of N (N3) are marked. Comparison of the appearance of dimeric MLONG and MCOMPACT (I).
