Abstract
Rotavirus is a major cause of severe acute gastroenteritis in the infants and young children. The past decade has evidenced the role of intrinsically disordered proteins/regions (IDPs)/(IDPRs) in viral and other diseases. In general, (IDPs)/(IDPRs) are considered as dynamic conformational ensembles that devoid of a specific 3D structure, being associated with various important biological phenomena. Viruses utilize IDPs/IDPRs to survive in harsh environments, to evade the host immune system, and to highjack and manipulate host cellular proteins. The role of IDPs/IDPRs in Rotavirus biology and pathogenicity are not assessed so far, therefore, we have designed this study to deeply look at the penetrance of intrinsic disorder in rotavirus proteome consisting 12 proteins encoded by 11 segments of viral genome. Also, for all human rotaviral proteins, we have deciphered molecular recognition features (MoRFs), which are disorder based binding sites in proteins. Our study shows the wide spread of intrinsic disorder in several rotavirus proteins, primarily the nonstructural proteins NSP3, NSP4, and NSP5 that are involved in viral replication, translation, viroplasm formation and/or maturation. This study may serve as a primer for understanding the role of IDPs/MoRFs in rotavirus biology, design of alternative therapeutic strategies, and development of disorder-based drugs.
Keywords: Rotavirus, Intrinsically Disordered Proteins (IDPs), Reoviridae, Diarrhea, MoRF
1. Introduction
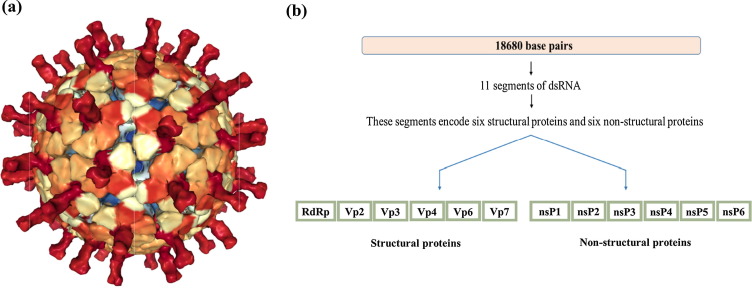
Diarrheal diseases are a major cause of morbidity and mortality in children aged below 5 years, predominantly in low-income nations [1]. According to WHO reports, diarrheal diseases cause more than 500,000 deaths per year worldwide. Rotaviruses belong to Reoviridae family and are documented as the primary causative agents of acute viral diarrhea in humans, animals, and avian species [2]. Among numerous serotypes of rotavirus, only five, such as G1, G2, G3, G4, and G9, are highly infectious. G3 serotype of rotavirus was highly infectious during 1994–1995 [3]. In 1995–1996, an outbreak of G9 serotype of rotavirus occurred, and this serotype was involved in more than 50% of cases of rotavirus infections during that period. From 1996 to 1999, most of the rotaviral infections were caused by G1 and G2 serotypes, with both serotypes being accountable for nearly 50% infections [3], [4]. Rotavirus is a non-enveloped RNA virus with a complex concentric triple-layered capsid that encloses the genome of 11 segments of double-stranded RNA (dsRNA) [5]. Each RNA segment codes for a single protein but the 11th segment encodes two non-structural proteins (NSP5 and NSP6) from overlapping open reading frames. As a result, the 11 segments of the viral genome encode 12 proteins, which are six non-structural proteins (NSP1, NSP2, NSP3, NSP4, NSP5, and NSP6) and six structural viral proteins (VP1, VP2, VP3, VP4, VP6, and VP7) [5] (Fig. 1 ).
Fig. 1.
(a) Atomic model of Rotavirus taken from PDB ID: 4V7Q [6](b) Schematic representation of all structural and non-structural proteins encoded by rotavirus genome segments. Rotavirus genomic dsRNA 18,680 base pairs (top bar, light pink color), encodes six structural (green color bar) and six non-structural proteins (light orange color bar).
The structural proteins of the virion govern cell entry, host specificity, antigenic specificities and enzymatic functions associated with viral genome replication and transcription. The non-structural proteins play vital roles in genome replication and evasion of innate immune response of the host [5]. Cryo-electron microscopy structure (PDB ID: 4V7Q) of rotavirus reveals the architectural organization of the triple-layered concentric capsid that surrounds the segmented RNA genome [6]. The diameter of mature rotavirion, which is characterized by the T = 13 icosahedral symmetry, is ~1,000 Å. The outer shell contains multiple copies of two proteins, VP4 & VP7, whereas the intermediate and inner capsids consist of VP6 and VP2, respectively. The outer surface of rotavirus exhibits 120 Å-long 60 spikes composed of VP4. The capsid features 132 aqueous channels with ~140 Å in length, spanning two outer capsid layers [7]. During virus entry, a sequence of molecular transformations in the outer layer involving the spike protein VP4 facilitates the internalization of the virus and release of the ‘double-layered particle’ (DLP) in the cytoplasm with the removal of the outer capsid [6]. Removal of the outer layer results in activation of the transcriptase and initiates synthesis and extrusion of the capped mRNAs into the cytosol [5], [6].
Although crystallographic structures and functions of most of the rotavirus proteins are known rather well, the intrinsically disordered aspects of these proteins have not been evaluated as of yet. Also, the crystallography provides only the static view of a protein inside crystal packing where disordered regions may undergo disorder to order transitions that ultimately hide the native flexibility and disordered propensity [8]. Therefore, our prime focus is to fill this gap by evaluating the natural content of intrinsic protein disorder inside rotavirus proteome. In general, disordered proteins (which are proteins structural features cannot be determined by the traditional structure determination methods, such as electron cryo-microscopy and X-ray crystallography) are represented by ‘Intrinsically Disordered Proteins’ (IDPs) and/or proteins containing ‘Intrinsically Disordered Protein Regions’ (IDPRs) that devoid of specific 3D structure in their native, biologically active forms [9], [10], [11]. IDPs/IDPRs have numerous crucial functions, being involved in the interaction with multiple partners and regulating many cellular processes. Recent studies implicated the direct role of IDPs/IDPRs in various human diseases, such as cancer, amyloidosis, diabetes, cardiovascular diseases, viral diseases, and many more [11]. In general, viruses, due to their limited protein-coding capacity, could gain several functional advantages from IDPs/IDPRs in terms of assembly of replication complexes involving a large number of viral and host proteins and survival in extreme host environments [12]. In recent studies, we have shown the prevalence of IDPs/IDPRs and their functional significance in Zika virus (ZIKV) and Chikungunya virus (CHIKV) [13], [14], as well as in Hepatitis C virus (HCV) [15], human immunodeficiency virus-1 (HIV-1) [16], various human papillomaviruses (HPVs) [17], respiratory syncytial virus (RSV) [18], Dengue virus [19], MERS-CoV [20], and Alkhurma virus (ALKV) [21].
IDPs/IDPRs are promiscuous binders in their nature, as they can have numerous interactions with distinct partners at a time, and can form important protein-protein interaction network hubs to control several signaling pathways at the same time [22]. In recent studies, it is reported that several IDPs/IDPRs display disorder-to-order transitions after interaction with their partners [23]. For example, the intrinsically disordered transactivation domain of c-Myb attains an α-helical conformation after association with KIX [24], [25], [26], [27], whereas C-terminal regulatory domain of p53 oncoprotein undergoes transitions from its disordered form into α-helical, β-structural, or two irregular forms with different configurations at binding to Cyclin A, Sirtuin, CBP, and the S100ββ dimer [23], [28]. To enable such folding at binding interactions with their partners, many IDPs/IDPRs are equipped with molecular recognition features (MoRFs) [29], [30], which are short disorder-based binding regions that regulate the functional mechanism under physiological conditions. Generally, viral proteins such as those of Zika, HPV, and Chikungunya virus have a higher frequency of MoRF regions, where MoRFs are identified as important factors in regulating the functional protein-protein interactions, having implications in drug discovery [31], [32], [33], [34].
In this work, we have examined the prevalence of intrinsic disorder in SA11 rotavirus proteome via multiple predictors and also performed PONDR® VSL2-based comparative intrinsic disorder analysis of different rotavirus serotypes (G1, G2, G3, G4, & G9). We have also predicted several MoRF regions in non-structural proteins of SA11 strain. We found a higher percentage of disordered regions in non-structural proteins, suggesting that their IDPRs might be involved in the regulation of assembly of replication complexes and processes and maturation of virus particles. This study could provide the important regions of rotavirus proteome that could be used as improtant targets for designing disordered-based small molecule inhibitors to block virus-host protein interactions for alternate therapy against the viral disease.
2. Materials and methods
We obtained all the reviewed protein sequences of simian rotavirus strain SA11 from UniProt which is most studied reference strain of rotavirus [35]. It has been widely used for experimental characterization of rotaviral pathogenesis globally [36], [37]. Therefore this strain seems to be most appropriate for the intrinsic disorder MoRF analysis. Furthermore, we have also obtained the reviewed protein sequences of most common human pathogenic rotavirus serotypes from UniProt to examine the comparative penetrance of intrinsic disorder. The list below provides the UniProt IDs for different proteins of these rotaviral serotypes: for G1 serotype, the UniProt IDs for NSP1-VP7 are (B3SRS0, B3SRS2, B3SRS1, Q9YJN7, B3SRS5, B3SRS6, B1NKR3, B1NKR4, B1NKR5, B3SRR9, B1NKR6, and P11853 respectively); for G2 serotype, the UniProt IDs for NSP1-VP7 are (P35423, Q03240, A1YTU9, B3SRT2, P23048, B3SRT4, A7J3A6, B1NKR8, Q6WNW5, P11196, A7J3A8, and P11850 respectively); for G3 serotype, the UniProt IDs for NSP1-VP7 are (B3SRV2, B3SRV4, B3SRV3, B3SRV6, B3SRV7, B3SRV8, B1NKS9, B1NKT0, B1NKT1, P11195, B1NKT2, and P11854 respectively); for G4 serotype, the UniProt IDs for NSP1-VP7 are (Q82045, B3SRX0, Q82053, Q82035, B3SRX3, B3SRX4, B1NKT7, B1NKT8, B1NKT9, P11200, B1NKU0, and P10501 respectively); and for G9 serotype, the UniProt IDs for NSP1-VP7 are (B3SRX6, B3SRX8, Q82049, B3SRY0, B3SRY1, B3SRY2, B1NKU1, B1NKU2, B1NKU3, B3SRX5, B1NKU4, and B3SRX9 respectively).
For intrinsically disordered regions (IDPRs) and MoRF analysis, numerous technologically advanced predictors are available, such as IUPred [38], ANCHOR [39], MoRFPred [40], MoRFchibi_web [41], DisEMBL [42], GlobPlot [43], DisoPred [44], PONDR® pool [including predictors, such as PONDR® VLXT [45], PONDR® VLS2 [46], and PONDR® VL3 [47], as well as a meta-predictor PONDR® FIT [48], and several others [49]. Critical Assessment of Protein Structure Prediction (CASP) was used to examine the accuracy of several disorder predictors, and several of the software mentioned above were compared using CASP [50]. All the available predictors are developed to identify the presence of intrinsically disordered residues and regions in query proteins, yet they all have diverse perspectives to examine the occurrence of intrinsic disorder. Therefore, it is sensible to use several computational tools to comprehend the abundance of intrinsic disorder in proteins [11]. All the predictors have limited accuracies of the prediction of intrinsic disorder in proteins, and the predictors whose accuracies are above 75% are considered as precise predictors. Characteristically, if the mean disordered score of any protein/region is above the threshold of 0.5, such proteins/regions are considered as IDPs/IDPRs. In addition, the iso-electric point of proteins has also been predicted by putting sequence in ExPasy (ProtParam tool) server [51].
Based on that standard, we selected four predictors from the PONDR family [PONDR® VLXT, [45] PONDR® VLS2, [46] PONDR® VL3, [47], PONDR® FIT [48]] and one more widely used server IUPred to analyze the prevalence of intrinsic disorder in rotavirus proteome. A meta-predictor PONDR® VL-XT integrates three predictors, the VL1 predictor trained to recognize long IDPRs, the amino-terminus-based predictor (XN), and the carboxyl-terminus-based predictor (XC) both trained to identify short disordered regions based on the X-ray crystallographic studies [45]. PONDR® VSL2 predictor uses a linear support vector machine (SVM) to predict both long and short IDPRs with 81% accuracy [46], [52]. PONDR® VL3 is trained for sequence-based predictions of long IDPRs (>30 amino acid residues) by calculating the means of 10 artificial neural networks and choose the final prediction [47]. A meta-predictor PONDR® FIT represents a combination of PONDR® VLXT, PONDR® VSL2, PONDR® VL3, FoldIndex, IUPred, and TopIDP that gives considerably enhanced precision in the collective as related to its different constituent predictors [48]. For each protein, we used the outputs of these five disorder predictors to calculate the corresponding PPID (predicted percent of intrinsic disorder) values, which correspond to the percent of residues of a query protein that were predicted to be disordered by a given predictor. We also calculated the mean PPID values for query proteins by averaging the outputs of all five predictors.
Apart from IDP analysis, we have also performed the MoRF predictions for each protein of SA11 rotavirus strain. In general, MoRFs are the short (5–25 residues) protein binding sites within disordered region sometimes longer also (more than 50 residues) and may have the potential to undergo disorder to order transitions upon interaction with other partners [53]. In our study, four standard MoRF predictors were used as: ANCHOR [39], MoRFPred [40], MoRFchibi_web [41] and Disopred [54]. All these predictors utilize different types of algorithms to predict MoRFs, so we have used four predictors to completely characterized these regions in rotavirus proteome. ANCHOR based predictions utilize estimated energy calculations relative to biophysical properties whereas MoRFpred identifies on the basis of sequence alignment and support vector machine (SVM) [39], [40]. Disopred provide disorder region based upon sequence property training sets trained through neural networks [54]. MoRFchibi_web is the most advanced and accurate predictor available; it gives the combined output of MoRFchibi and MoRFDC by following the Bayes rules [55]. Therefore, we have used only MoRFchibi_web-based predictions to map the regions on available crystal structures of rotavirus proteome.
We have also investigated the multiple sequence alignment of G1, G2, G3, G4 and G9 serotypes with the SA11 strain of rotavirus by Clustal Omega, web-based server [56]. All sequences were analyzed on the basis of three annotations which includes, conserved positions, strong polymorphisms and weak polymorphisms. The conserved regions have similar amino acid sequence throughout all strains. The strong polymorphisms are represented by space in the output file, which means substitutions among strongly dissimilar amino acid residues according to PAM 250 matrix. The weak polymorphisms are regions which have conserved residues among similar sequences based on their properties. These are denoted by colon and period symbols based on their scores having a threshold of 0.5 in PAM 250 substitution matrix.
The predicted MoRF regions were also checked for functional annotations by using eukaryotic linear motif (ELM) server [57] which gives the information of short linear motifs (SLiMs) (3–10 residues) within IDPRs having conserved functional sites. These motif sequences provide an interface for functional interaction with other binding partners with low binding affinity. In this study we have predicted functional sites in rotavirus proteins from strain SA11 using ELM (Eukaryotic Linear Motif) server, which constitutes the manually annotated instances (matched sequences) curated from experimental literature [58].
3. Results and discussion
3.1. Examination of intrinsic disorder status of rotavirus SA11 proteins
Proteins and protein regions that do not possess stable and precise three-dimensional (3D) structures under physiological conditions are known as IDPs/IDPRs. As explained before, during co-crystallization of proteins with their natural partners, interaction with said partners can stimulate binding-induced folding of IDPRs resulting in the appearance of specifically structured conformations, suggesting that the actual content of intrinsic disorder in proteins could be noticeably higher than the quantity projected from the missing electron density. Although the crystal structures of some of the rotavirus proteins are deposited in the Protein Data Bank (PDB), in the absence of complete structural knowledge pertaining to these proteins in solution, the use of computational means represents the best way to detect IDPRs in a given protein, thereby providing an important way of generating crucial information regrading protein structure and fucntion.
3.2. Examination of intrinsic disorder in rotavirus structural proteins
3.2.1. RNA-dependent RNA polymerase (RdRp/VP1)
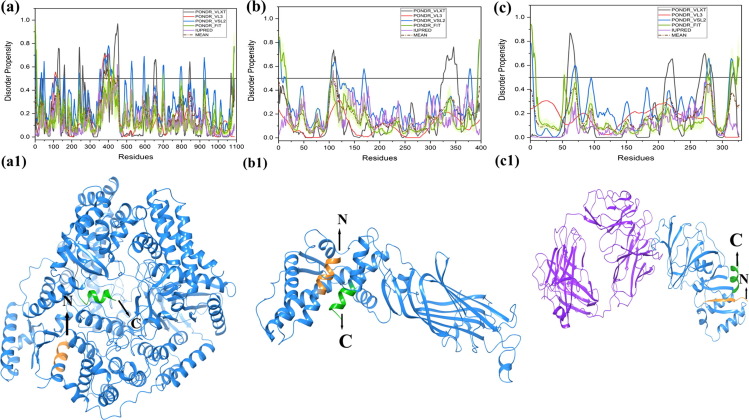
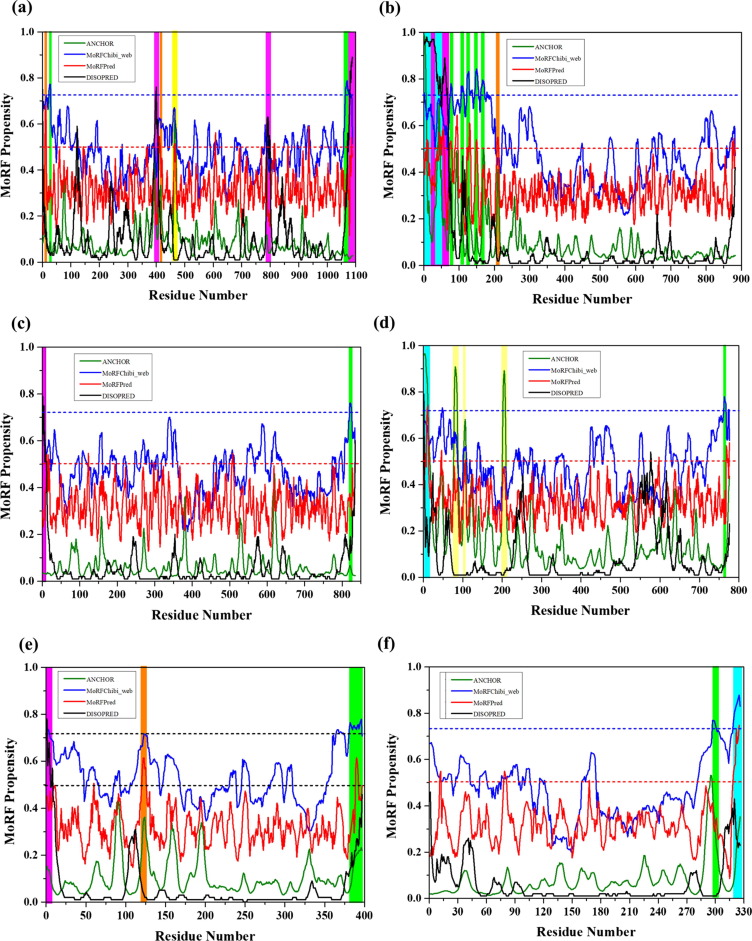
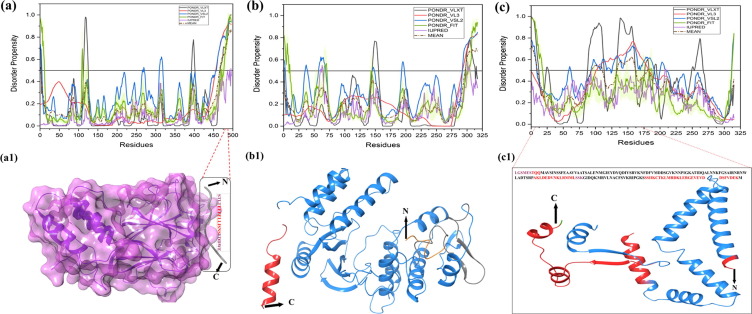
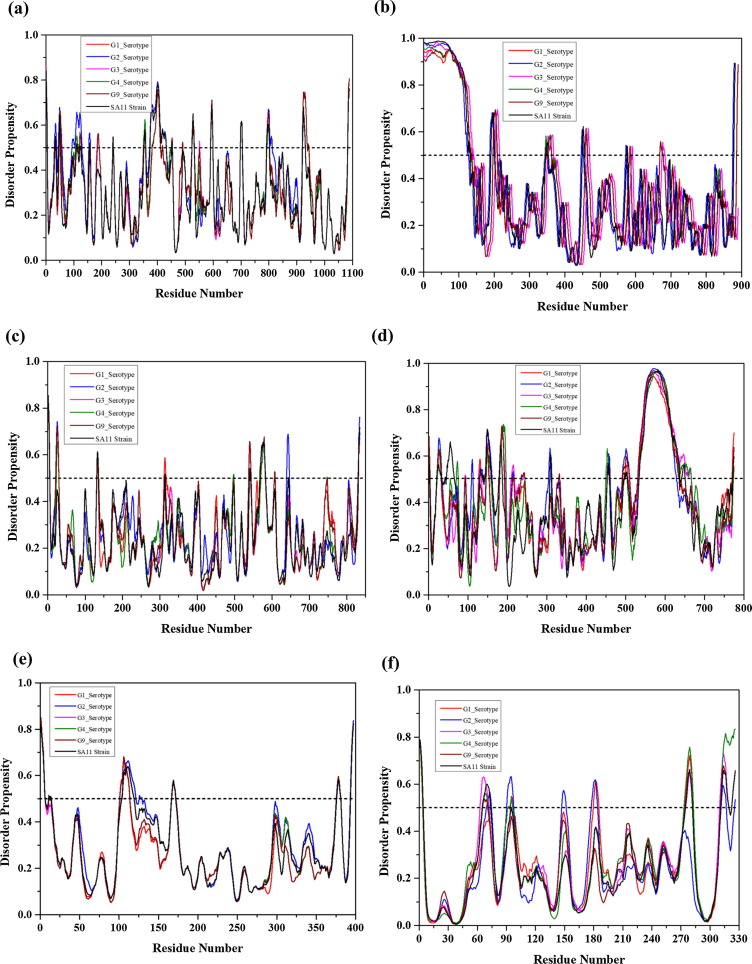
RdRp, also known as VP1, is encoded by the gene segment 1 in all rotavirus strains. It is a compact globular protein with a diameter of 70 Å. Its 3D crystallographic structure is known (PDB ID: 2R7Q), containing three domains, such as N-terminal domain (residues 1–332), polymerase domain comprising finger subdomains, palm subdomain, and thumb subdomain (residues 333–778) and a C-terminal domain (residues 779–1089) [59]. A priming loop (residues 557–567), also known as a “primer grip,” situated at the joining site of thumb and palm subdomains, has a flexible structure that interacts with triphosphates during initiation. The catalytic activity of VP1 is enhanced by the interaction between the residue Leu138 of NSP1 protein with NSP2/NSP5 non-structural proteins under physiological condition [60]. Our analysis revealed that the mean PPID of the RdRp protein is rather low (3.1%) { Table 1 }, this protein is having compact globular structure with few predicted MoRFs in its polymerase domain (residues 393–407 and 463–471) that may be responsible for the polymerase activity {Fig. 2.1(a) and Fig. 2.1 (a1)} {Table 2 and Fig. 3 (a)}. Since no MoRFs have been mapped in predicted disordered region (residues 390–404) therefore we have not given the functional annotation of MoRFs through ELM server for VP1 (Table S1).
Table 1.
Analysis of intrinsically disordered properties of SA11 rotavirus proteins.
| Protein | PPIDVLXT | PPIDVSL2 | PPIDVL3 | PPIDFIT | PPIDIUPRED | PPIDMEAN |
|---|---|---|---|---|---|---|
| RdRp/VP1 | 11 | 13.4 | 6.6 | 2.2 | 0.4 | 3.1 |
| VP2 | 17.4 | 18.5 | 14.9 | 11.9 | 7.7 | 14.2 |
| VP3 | 4.6 | 4.2 | 0 | 2.0 | 0 | 0 |
| VP4 | 4 | 11.2 | 0 | 15.7 | 6.4 | 12.8 |
| VP6 | 10.3 | 10.3 | 0 | 4.8 | 0.5 | 0.5 |
| VP7 | 15.3 | 9.2 | 0 | 7.1 | 0 | 0 |
| NSP1 | 10.7 | 11.1 | 8.3 | 6.9 | 0.4 | 6.7 |
| NSP2 | 9.2 | 15.8 | 7.9 | 9.1 | 2.2 | 6.6 |
| NSP3 | 30.2 | 36.8 | 25.7 | 7.9 | 0.6 | 17.5 |
| NSP4 | 39.4 | 51.4 | 45.7 | 22.3 | 9.1 | 32 |
| NSP5 | 54 | 96.5 | 100 | 86.9 | 56.4 | 84.3 |
| NSP6 | 6.5 | 10.9 | 0 | 15.2 | 0 | 4.3 |
Fig. 2.1.
Schematic illustration of the structural and intrinsic disorder characteristics of rotaviral structural proteins. The outpus of PONDR® VL-XT, PONDR® VL3, PONDR® VSL2, PONDR® FIT, and IUPred predictors are shown by black, red, blue, olive, and violet colors, respectively, whereas short dashed lines of wine color represent the mean disorder. Light olive shadow around PONDR® FIT curves represents the error distribution. The Morf regions is shown in grey color in the RdRp protein structure (a1). Plots signify disorder status of (a) 1,088 residue-long RdRp/VP1 that contains several IDPRs at its N-terminus; (b) 397 residue-long VP6 that possesses several IDPRs at its N- and C-terminal regions; (c) 326 residue-long VP7 is characterized by the presence of an IDPR at its N-tail and several IDPRs within the C-terminal domain. X-ray crystal structres are shown for RdRp protein (PDB ID: 2R7Q) (a1), VP6 protein (PDB ID: 1QHD) (b1), and VP7 protein (PDB ID: 3FMG) is structurally determined in complex with light and heavy chains of Fab of neutralizing antibody 4F8 (c1). The N and C-terminals are denoted in orange and green colors with arrows respectively. Sequence of experimentally determined structure is shown (disordered residues in red color). Some residues are missing in the structures that are shown in plum color in the sequence.
Table 2.
MoRF analysis of all proteins of SA11 rotavirus strain by four different tools. Red numeric values show the MoRF regions that are common between two or more predictors.
 |
*We have shown only the predicted disordered regions of each protein where MoRF residues are lying within that region.
Fig. 3.
Evaluation of disorder-based interactivity of the rotavirus structural proteins. The presence of MoRF regions was evaluated by ANCHOR (olive lines), DISOPRED (black line), MoRFpred (red line) and MoRFchibi_web blue lines). The threshold for MoRF predictions by ANCHOR, DISOPRED, MoRFPred are 0.5 and of MoRFchibi_web is 0.725. These thresholds are shown as dashed red lines and blue line, respectively. Positions of MoRFs predicted by ANCHOR is shown by a yellow bar, orange bar by MoRFPred, pink bar by DISOPRED, overlapped common MoRF positions are shown by cyan bar, and MoRFchibi_web are shown by olive bars, respectively. Plots signify MoRF analysis of (a) VP1 (b) VP2 (c) VP3 (d) VP4 (e) VP6 (f) VP7.
3.2.2. Structural protein VP2
The structural protein VP2, encoded by gene segment 2, forms the innermost capsid of the triple-layered viral shell. This layer encapsidates the VP1 and VP3 structural proteins in the vicinity of the 5-fold axes, covering the viral genome, and RNA processing enzymes [61]. It is not only anchored with VP1, but the 5-fold axes of VP2 are also an indispensable cofactor for the initiation of genome replication [62]. T = 1 symmetry of VP2 is needed for the initiation of the process of dsRNA synthesis [63]. Although the function of the N-terminal region (residues 1–132) is not understood well, this region seems to be important for the activation/localization of the RNA-dependent RNA polymerase (VP1) and guanylyl transferase-methylase (VP3) into the virion center [64]. Our intrinsic disorder predisposition analysis revealed that the mean PPID of VP2 is 14.2% {Table 1}, indicating that it can be classified as moderately disordered protein. This analysis also showed that the N-terminal region of the VP2 protein is predicted to be completely disordered (residues 1–120) {Fig. 2.1((a)} and also possess MoRF regions predicted by several algorithms {Table 2 and Fig. 3(b)}. For example, MoRFchibi_web has predicted 5 MORF regions. Experimental studies showed that the deletion of first 25 N-terminal residues of VP2 (a region that is predicted by ANCHOR and DISOPRED to contain MoRFs) entirely prevents the RNA binding activity and incorporation of VP1 and VP3 into virus-like particles (VLPs) [64]. The principal domain of VP2 (residues ~ 100–880) associated with the RdRp polymerase activity also exhibits several IDPRs (residues 100–120, 140–144, and 192–200) {Fig. 2.2((a)}, which have clear functional implications in virus maturation [61]. Also, the MoRFs present in these disordered regions indicates the essential disordered based binding sites are regulating the overall function and also predicted their functional annotations with ELM server (Table S1).
Fig. 2.2.
Schematic illustration of the structural and intrinsic disorder characteristics of rotaviral structural proteins. (a) 822 residue-long VP2 that includes long IDPR at its N-terminal region; (b) 835 residue-long VP3 that shows least level of disorder with no IDPRs; (c) 755 residue-long VP4 consisting of VP8* (residues 1–231) and VP5* (residues 248–776) that displays higher disorder levels at its C-terminus. The color schemes are same as Fig. 2.1.
3.2.3. Structural protein VP3
Structural protein VP3 is encoded by genome segment 3. It is a multi-functional capping enzyme of 835 amino acids [65]. While the N-terminal region of ~690 residues contains domains needed for mRNA capping functions (a variable N-terminal domain, a guanine-N7-methyltransferase (N7-MTase) domain, a ribose-2′-O-methyltransferase domain, a guanylyltransferase and RNA 5′-triphosphatase (GTPase/RTPase) domain), the C-terminal region of ~150 amino acids possesses a 2′,5′-phosphodiesterase (PDE) activity that cleaves 2′-5′-oligoadenylate [66], [67], thereby preventing RNase L activation [66], [67]. Apart from capping-associated activities, VP3 is also associated with virulence. In its N7-MTase and GTase/RTPase domains, VP3 has some sequence motifs that are conserved among the rotavirus and orbivirus species. The conserved residues in the N7-MTase domain are located in region 134–139, whereas in GTase/RTPase domain, residues from 552 to 555 and Ser527, Arg531, Trp571, His610 are highly conserved [66], [67]. Even though this multifunctional protein has a mean PPID score of 0% {Table 1}, individual predictors, such as PONDR® VLXT, PONDR® VSL2, and PONDR® FIT have predicted short stretches of disorder at residues 179–184, 242–246, 335–355, 814–819 (aa residues), 1–5, 131–135, 313–314, 540–542, 568–582, 644, and 832–835 (aa residues) and 1–8 and 827–835 (aa residues), respectively while IUPred has not predicted any region as disordered. However, two MoRF regions {Table 2 and Fig. 3(c)} were observed with functional annotations given by ELM server (Table S1) seems important for the functionality of this protein {Fig. 2.2((b)}.
3.2.4. Structural protein VP4
VP4, a non-glycosylated outer capsid spike protein, is encoded by the 4th genome segment. The trypsin-mediated proteolytic cleavage of VP4 into the N-terminal VP8 (molecular weight ~28,000 Da) and C-terminal VP5 (molecular weight ~60,000 Da) fragments enhance the virus infectivity [68]. The negatively charged VP4 protein comprises several random coils and turns in its N-terminal region [69]. This N-terminal region possesses MoRF forming residue {Table 2 and Fig. 3(d)}. Four conserved cysteine residues; i.e., Cys203, Cys216, Cys318, and Cys380, are present in all the rotavirus strains [70]. Trypsin cleavage at Arg241 and Arg247 yields the receptor binding VP8* fragment and VP5* fragment, that enhances the viral infectivity [70], [71], [72]. The 265–474 region of VP5* possesses membrane permeabilization property and is highly disordered as predicted by all individual predictors {Fig. 2.2((c)} [73], [74] VP4 is known to undergo structural rearrangements during membrane penetration [73]. Overall, VP4 is associated with multiple functions, such as cell surface interaction, neutralization, penetration, virulence, host range, and hemagglutination [73], [74], [75], [76], [77]. VP4 also binds microtubules and actin [78], [79]. The mean PPID score for VP4 is only 12.8% {Table 1} but the individual predictors PONDR® VLXT, PONDR® VL3, PONDR® VSL2, PONDR® FIT, and IUPred have predicted higher percentage of disordered regions in total 157, 97, 209, 123, and 50 residues respectively {Fig. 2.2(c)}. The N-terminal and C-terminal regions of this protein exhibit intrinsic disorder and MoRF sites that help in spike formation at the outer layer of the virus particle [6].
3.2.5. Structural protein VP6
The intermediate capsid protein VP6 is encoded by the sixth gene segment and serves as the major structural protein of the virion [5]. It consists of 397 residues that are highly conserved among group A rotavirus strains. VP6 determines the group and subgroup antigenic specificities of the virus [80]. The protein has a well-defined 3D structure (PDB ID: 1QHD) {Fig. 2.1(b1)} that self assembles into a trimer with two different domains: a β-barrel domain, situated away to the center and a proximal α-helix domain, positioned closer to the center. VP6 plays a critical role in the organization/maturation of the virion by interacting with an assembly of the inner and outer capsid layers, and packaging of the genomic RNA in the inner capsid [7]. The region between amino acids 105 and 328 near the center of the protein mediates Trimerization, and the C-terminal region between residues 251 and 397 mediates association of the protein with VP2 of the single-shelled particle [81]. Antibodies against VP6 inhibit viral mRNA synthesis, suggesting a role for the protein in mRNA synthesis within the DLPs [82]. Though this protein has a crystal structure and a mean PPID value of 0.5% {Table 1}, the N-terminal region is predicted to be intrinsically disordered by the individual predictors, PONDR® VLXT (residues 103–115), PONDR® VSL2 (residues 1–6, 11–14, and 104–119 residues), and PONDR® FIT (residues 1–15) while IUPred has predicted only two residues 107 and 108 to be disordered. {Fig. 2.1 and 2.1(b)}. Three MoRF predictors have shown only single MoRF region in VP6 protein (Table2, Fig. 3e). MoRFchibi_web predicted MoRF residues at C-terminal (381–396) have shown to play a crucial role in the maturation and organization of the virus particle [81].
3.2.6. Structural protein VP7
VP7 is the second most abundant capsid protein of the virion and is encoded by 9th gene segment of the rotavirus strain SA11. Both VP7 and VP4 are highly immunogenic and induce neutralizing antibodies, and VP7 exhibits several antigenic epitopes [83], [84]. The mean PPID value of this protein is (0%) {Table 1}, and VP7 has a well-defined crystal structure (PDB ID: 3FMG) {Fig. 2.1(c1)} that is enriched in α-helices and β-sheets. VP7 contributes the stability of the VP4 spikes at the outer surface of the virion and in the process of infection [83], [84]. Although it does not show any predicted disordered regions, the crystallographic conformation of VP7 demonstrates that the N-terminal arm (residues 58–78) defines a grip that forms a connection between VP7 and VP6, and this region is disordered in nature [83], [85]. Two of the five predictors {PONDR® VLXT (residues 58–71) and PONDR® VSL2 (residues 66–73)} suggest that these regions are disordered in nature. The C-terminal region (residues 313–325) of the VP7 protein is also disordered in nature and also has MoRF regions that may associate with the local dyad within its trimer {Table 2 and Fig. 3(f)}. The ELM server has predicted many SLiMs for VP7 and the MoRF residues have been functionally annotated in these SliMs (Table S1). The same residues are predicted to be disordered by the individual predictors, PONDR® VLXT (residues 317–319), PONDR® VSL2 (residues 312–319 and 323–326), and PONDR® FIT (residues 313–319 and 323–326). In crystal structure of VP7 (residues 50–330), predicted disorder residues (310–328) are missing and hence are in agreement with our disorder predictions. Experimental studies have suggested a role for the disordered N- and C-terminal domains in the organization and maturation of the virions by undergoing significant conformational changes, where the aqueous interface of inner surface of VP7 allows protein interaction with the outer surface of VP6 by one pair of van der Waals interaction (Pro279 and Thr281 of VP7 interacting with Pro313 of VP6) and one side chain H-bond (Gln305-Asn310) [83].
3.3. Exploration of the intrinsic disorder status of the rotaviral non-structural proteins
3.3.1. Non-structural protein NSP1
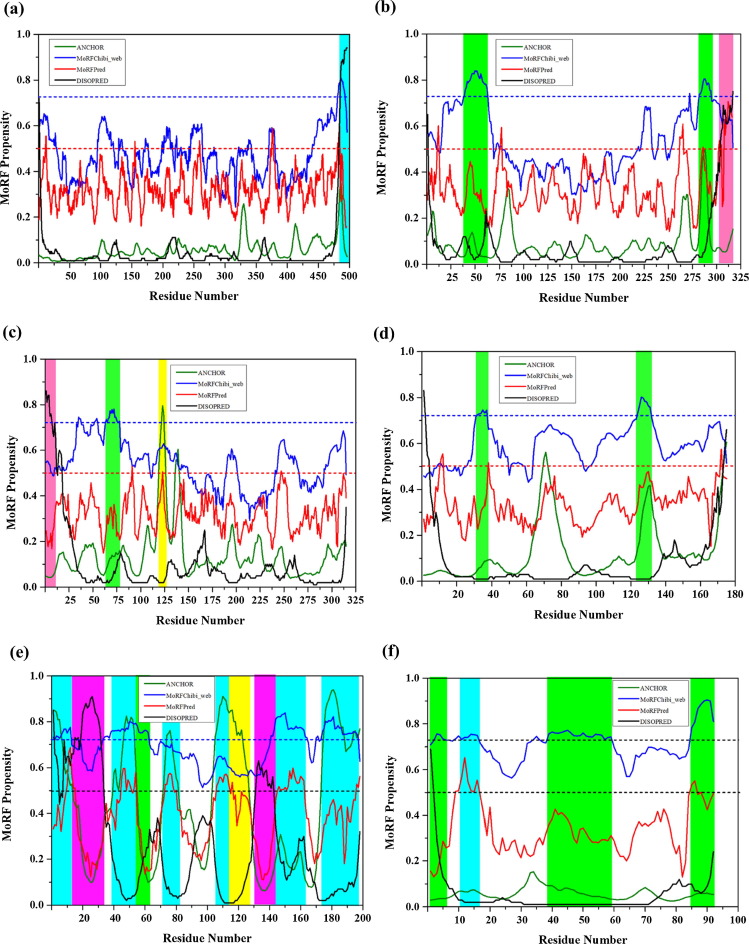
NSP1 is a non-structural RNA binding protein, encoded by the 5th genome segment. It is a basic protein and the most divergent among the rotaviral proteins. The N-terminal region of 150 residues is relatively more conserved than the rest of the protein and contains three basic regions at residues 10–39, 79–91 and 111–126 and a conserved cysteine-rich motif between residues 42 and 75 that forms a zinc-finger. A second non-conserved Cys-rich motif is also present between residues 314 and 327 [86], [87], [88]. Mutant viruses containing the NSP1 gene coding for only the amino-terminal 50 amino acids are able to replicate, suggesting that the region downstream of residue 50 not be essential for virus replication [88]. The partial crystal structure of NSP1 has been determined (PDB ID: 5JER) that has C-terminal stretch (475–500) bound to IRF3 protein {Fig. 4.1(a1)}. Analysis of the complete sequence of NSP1 protein (1–495 residues long) using the five different predictors showed the mean PPID score of 6.7% {Table 1}. PONDR® VSL2 predicted disordered regions (311–319 residues) that comes under zinc-binding site (314–327 residues) {Fig. 4.1 (a)}. Overall the C-terminal region is predicted to be disordered by all the predictors {Fig. 4.1(a1)} containing MoRFs precited by MoRFchibi_web and DISOPRED (residues 481–492 and, 483–497) {Table 2 and Fig. 5(a)}. The ELM server predictions have shown many binding sites for MoRF regions of NSP1 as shown in supplementary file (Table S1).
Fig. 4.1.
Graphical illustration of the structural and intrinsic disorder characteristics of rotaviral structural proteins. The outputs of PONDR® VL-XT, PONDR® VL3, PONDR® VSL2, PONDR® FIT and IUPred predictors are shown by black, red, blue, olive, and violet colors, respectively, whereas short dashed lines of wine color represent the mean disorder. Light olive shadow around PONDR® FIT curves represents the error distribution. The red color indicates the mean disorder region present in the protein structures and the grey color represents the MoRF regions present in NSP1 (a1), NSP2 (b1), and NSP3 (c1). The N and C-terminals are denoted in orange and green colors with arrows respectively. Plots signify the disorder status of NSP1 (a); NSP2 (b); and NSP3 (c). X-ray crystal structures are shown for NSP1 protein (PDB ID: 5JER) in complex with Interferon regulatory factor 3 (IRF3, shown in violet color) (a1); NSP2 (PDB ID: 6CY9) (b1); and NSP3 protein (PDB ID: 1KNZ) (c1).
Fig. 5.
Evaluation of disorder binding regions of the rotavirus non-structural proteins. The presence of MoRF regions was evaluated by ANCHOR (olive lines), DISOPRED (black line), MoRFpred (red line) and MoRFchibi_web blue lines). The threshold for MoRF predictions by ANCHOR, DISOPRED, MoRFPred are 0.5 and of MoRFchibi_web is 0.725. These thresholds are shown as dashed red lines and blue line, respectively. Positions of MoRFs predicted by ANCHOR is shown by a yellow bar, orange bar by MoRFPred, pink bar by DISOPRED, overlapped common MoRF positions are shown by cyan bar and MoRFchibi_web are shown by olive bars, respectively. Plots signify MoRF analysis of (a) NSP1 (b) NSP2 (c) NSP3 (d) NSP4 (e) NSP5 (f) NSP6.
3.3.2. Non-structural protein NSP2
NSP2 is encoded by the 8th gene segment in SA11 strain of rotavirus. It is an essential nucleating protein of viroplasms, which function as viral factories, where viral genome replication and assembly of double-layered particles (DLPs) take place [89]. Its interaction with NSP5 leads to viroplasm formation [90]. NSP2 interacts with several host proteins and sequesters them into the viroplasm [91]. It also interacts with tubulin and is associated with the perinuclear localization of the viroplasms [92], [93]. Crystallographic structure (PDB ID: 6CY9) of NSP2 has revealed that it forms a large doughnut-shaped octamer through tail-to-tail tetramer assemblies {Fig. 4.1(b1)} [94]. It possess non-specific sequence-independent RNA-binding activity and helps in efficient viral mRNA replication in combination with the viral polymerase complex [89], [94], [95], [96]. NSP2 also possesses Mg2+-dependent NTPase, helix-unwinding, and NDP kinase activities [96], [97], [98]. Residues responsible for NDP kinase activity, such as Thr 56, His 59, Phe 64 are located within the MoRF region (38–61), which also includes some of the residues 53 to 76 that form part of RNA-binding grooves {Table 2 and Fig. 5(b)} This multifunctional protein showed a mean PPID value of about 6.6% {Table 1}. {Fig. 4.1(b1)} shows that the C-terminal RNA-binding domain (residues 295–317) is predicted to be mostly disordered, suggesting that it undergoes the conformational changes after interacting with the viral polymerase complex. Since this protein interacts with host proteins, our ELM predictions annotated several binding sites for NSP2 MoRFs as shown in supplementary file (Table S1).
3.3.3. Non-structural protein NSP3
The 7th genome segment encodes NSP3 protein [99]. The protein is slightly acidic and binds specifically the conserved tetranucleotide sequence present at the 3′ end of the rotaviral mRNAs [100]. There is a crystal structure available for NSP3 (PDB ID:1KNZ) {Fig. 4.1(c1)} with a dimerization region (residues 150–206), an eIF4G-binding region (residues 274–313) and an RNA binding domain (residues 83–149) [101], [102], [103]. The RNA-binding domain helps the protein to act as dimer in vivo and is implicated in facilitating viral mRNA translation by competing with and evicting the poly(A)-binding protein (PABP) from eIF4GI and shut-off of host cellular protein synthesis [104], [105], [106], [107]. The overall mean PPID score of NSP3 protein is 17.5% {Table 1} with higher disorder content being found within its dimerization region (residues 150–206), and in the RNA binding domain (residues 83–149) {Fig. 4.1(c)} that are important for the protein function. Also, the residues 150–160 are missing in PDB structure and are predicted to be disordered through mean PPID region {Fig. 4.1(c1)}. The RNA binding domain also comes under MoRF region predicted by ANCHOR {Table 2 and Fig. 5(c)}. Experimental studies have shown that mutations in the flexible region (four amino acid residues, W170A, K171E, R173E, and R187E: K191E) reduced the stable dimer formation and altered the protein stability [107]. In agreement with these observations, disorder prediction analysis has also shown that this is characterized by the enhanced disorder predisposition.
3.3.4. Non-structural protein NSP4
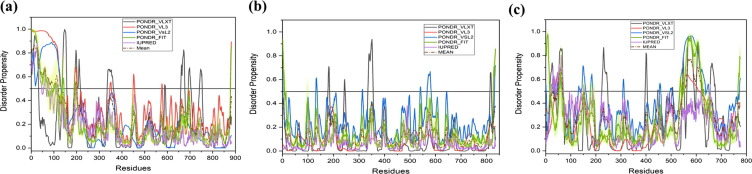
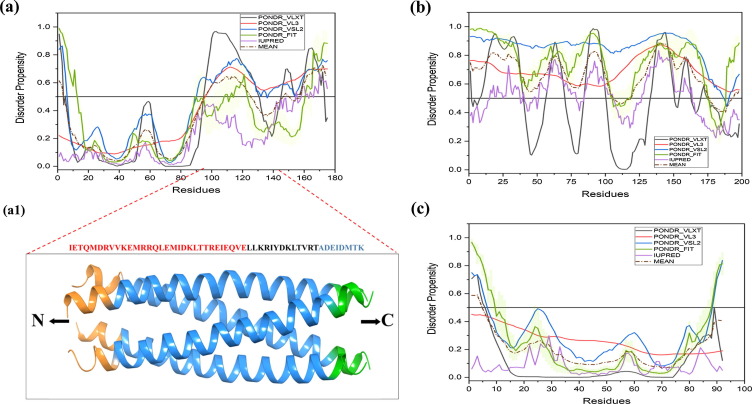
NSP4 (20.26 kDa) is a 175 residue-long glycosylated protein encoded by the 10th genome segment [108]. It is localized to the endoplasmic reticulum (ER) through its N-terminal hydrophobic regions and functions as the ER-anchored receptor for DLPs for their maturation into TLPs in the ER and is essential for the morphogenesis of the virion [108]. It is also localized in other organelles as well as is secreted from the apical surface of the polarized epithelial cells [109]. It is the first identified viral enterotoxin [110], [111]. The flexible C-terminal cytoplasmic region (residues 161–175) of NSP4 binds to VP6 of the DLPs and mediates their entry into the ER lumen where the virus becomes temporarily enveloped [112], [113] and matures into TLP. NSP4 interacts with microtubules and immobilizes the early secretory pathway [114], [115]. The NSP4 protein shows overall 32% mean PPID score {Table 1} with higher levels of disorder in cytoplasmic domain (residues 95–146), enterotoxic peptide region (residues 114–135), and the flexible C-terminal region (residues 139–175) {Fig. 4.2(a)}. Although, the region from residue 114 to 135 forms α-helical bundle in crystal structure {Fig. 4.2(a1)}, however it has been predicted to have high disorder propensity based on amino acid composition. This is in line with the known fact that under specific conditions used to promote protein crystallization process, disordered regions tend to form some secondary structure elements [116]. Also, in a study, NSP4 peptides (residue 120–147) has shown disordered type of CD spectra in buffer and undergo significant conformational transitions in membranous environment [117]. The aforementioned disordered regions of NSP4 play a key role in the initiation of diarrhea and binding to DLP [112], [113]. The MoRF region (residues 124–131) resides in the region that is responsible for many functions such as residue 112–148 utilized as the VP4 binding domain residues 112–140 for Caveolin binding, and residues 112–135 used for Ca+2 binding and enterotoxin domain {Table 2 and Fig. 5(d)}. Functional annotations with predicted SliMs have also been shown in supplementary file (Table S1). Amino acid substitution mutations P168A, Y166S, and M175I in the unstructured C-terminus of NSP4 lead to the complete loss of interaction of the protein with DLPs [113]. This multifunctional protein is pleiotropic in nature because of the oligomeric state combined with high conformational dynamics of the disordered cytoplasmic domain of the protein as evident from the calcium-lacking pentameric (PDB ID: 1G1J & 2O1K) [118], [119] and the calcium-containing tetrameric (PDB ID: 2O1J) [120], [121], [122] coiled-coil structures of the diarrhea-inducing region from ST3, and SA11 and I321 strains of rotavirus, respectively. NSP4 has a cholesterol recognition amino acid consensus sequence (CRAC), which is predicted to be a completely disordered region and it interacts with cholesterol and caveolin, leading to its localization to the plasma membrane [123].
Fig. 4.2.
Graphical illustration of the structural and intrinsic disorder characteristics of rotaviral structural proteins. Plots indicate the disorder status of NSP4 (a); NSP5 (b); and NSP6 (c). Crystallized structure of diarrhea-inducing domain of NSP4 (PDB ID: 2O1J, residues 95–140) is shown in (a1).
3.3.5. Non-structural protein NSP5
The 11th gene segment encodes NSP5 protein (21.70 kDa) [124]. It undergoes O-linked glycosylation [125] and phosphorylation [126], [127], and interaction with NSP2 leads to the hyperphosphorylation of NSP5 [90], [128]. NSP5 is crucial for viroplasm formation and recruitment of other viral proteins [129]. Both the N-terminal (residues 1–33) and C-terminal (residues 131–198) regions of the NSP5 are important for interaction with NSP2, viroplasm formation, and virus replication [130]. NSP5 protein of rotavirus has the highest intrinsic disorder level among all the rotaviral proteins with mean PPID of 84.3% {Table 1}. {Fig. 4.2(b)} shows that NSP5 is predicted to have long IDPRs in its N-terminal region, and within the C-terminal domain that contains the CXC motif (C171 and C174) responsible for the iron-sulfur cluster organization. There are various MoRF regions present in NSP5 protein of rotavirus SA11 and other strains that are involved in the functionality of potential protein kinase C phosphorylation sites (S-X-R/K-R/K-X-X-S and K/R-X-S) at serine residues 22, 30, 75, 100, and 136 [131] {Table 2 and Fig. 5(e)}. There are also serine residues at positions 56, 154, and 165 that are potential substrates of casein kinase II (S-X-X-D/E) {Table 2 and Fig. 5(e)}. Phosphorylation of Ser-67 within the SDSAS motif (amino acids 63–67) was required to trigger hyperphosphorylation by promoting the activation function, and mutation of all three Sers of motif a (Ser-63, Ser-65, and Ser-67) completely abolished activation function [132]{Table 2 and Fig. 5(e)}. Recent studies demonstrated cytoplasmic relocalization of several nuclear hnRNPs (heterogeneous nuclear ribonucleoproteins), and ARE-BPs (AU-rich element-binding proteins) during rotavirus infection, influencing viral gene expression/replication and progeny virus production [91]. Rotavirus was also demonstrated to induce atypical stress granules and P-bodies during infection [133]. These studies further demonstrated that majority of the relocalized nuclear proteins, some cytoplasmic proteins, as well as the atypical stress granules and P-bodies formed during rotavirus infection are sequestered in the viroplasms largely through the interaction of the host proteins with NSP2 and/or NSP5 in the viroplasm [91], [133]. Intrinsic disorder appears to be critical for the multifunctionality of NSP5. The presence of high intrinsic disorder level in NSP5 protein is expected based on the peculiarities of its genetic makeup. In fact, viruses have evolved a complex genetic organization for optimal use of their limited genomes and production of all necessary structural and regulatory proteins, including rather frequent utilization of overlapping open reading frames [12], [16], [134], [135]. It has been shown that proteins or protein fragments corresponding to overlapping genes are either disordered or possess complementary disorder distribution, where if a protein encoded by one ORF is ordered, then the protein encoded by the overlapped ORF will have high levels of intrinsic disorder [12], [16], [134], [135].
3.3.6. Non-structural protein NSP6
NSP6 (11.01 kDa) is another viral protein produced from an overlapping open reading frame (ORF) formed by the gene duplication of segment 11 [136]. Interestingly, the NSP6 proteins has varying length of amino acid residues in different rotavirus strains such as SA11 has NSP6 with 92 amino acid residues where in some strains NSP6 is truncated with less than 40 residues (G1, G3 and G9) [137]. In comparison to other non-structural proteins, NSP6 is expressed at a very low level and is observed to be unstable [138]. It binds DNA and RNA in a sequence-independent manner with equal affinity. The first 20 residues at the N-terminal region are essential for mitochondrial targeting and altering the mitochondrial function during rotavirus infection [139]. NSP6 is localized to viroplasm after phosphorylation and regulates self-association of NSP5 [138]. The protein is only slightly disordered in nature and has the mean PPID score of 4.3% {Table 1}. {Fig. 4.2(c)} demonstrates that NSP6 has two short, terminally-located IDPRs. The N-terminal disordered region (residues 1–6) also has MoRF forming residues (1–20) are essential for mitochondrial targeting and alter the mitochondrial function during rotavirus infection {Table 2 and Fig. 5(f)}. Recent reverse genetics experiments demonstrated that NSP6 is not essential for rotavirus replication in the cell culture [140].
3.4. Comparative analysis of intrinsic disorder status in proteins from different rotavirus serotypes
There are multiple different serotypes of rotavirus that cause the rotaviral infections worldwide, with the most causative serotypes being G1, G2, G3, G4, and G9, which are collectively responsible for more than 95% of rotavirus infections. The infection rate of these serotypes mainly dependent on the geography and temporal regions of the rotavirus (as shown in Table 3 ) [4].
Table 3.
The infection rate (in percentage) of different serotypes of rotavirus based on the geography and temporal regions.
| Regions | Serotypes |
||||
|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G9 | |
| South America | 34 | 23 | 2 | 9 | 15 |
| Africa | 23 | 2 | 21 | 4 | 7 |
| Asia | 34 | 13 | 1 | 20 | 12 |
| North America | 73 | 11 | 6 | 4 | 3 |
| Europe | 71.6 | 9 | 2 | 11 | 4 |
| Australia/Oceania | 82.4 | 14 | 1 | 2 | 0.5 |
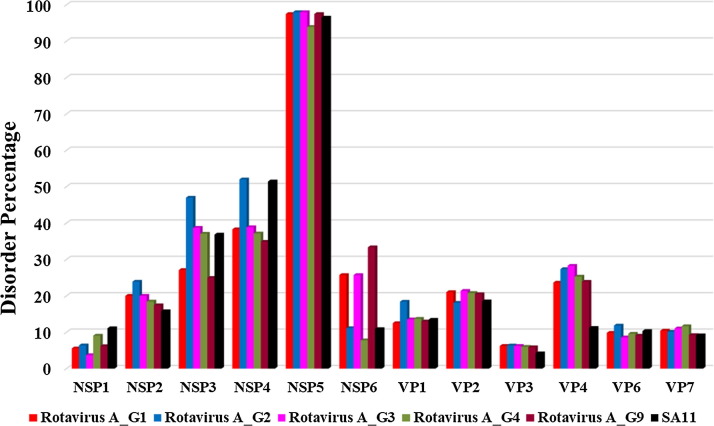
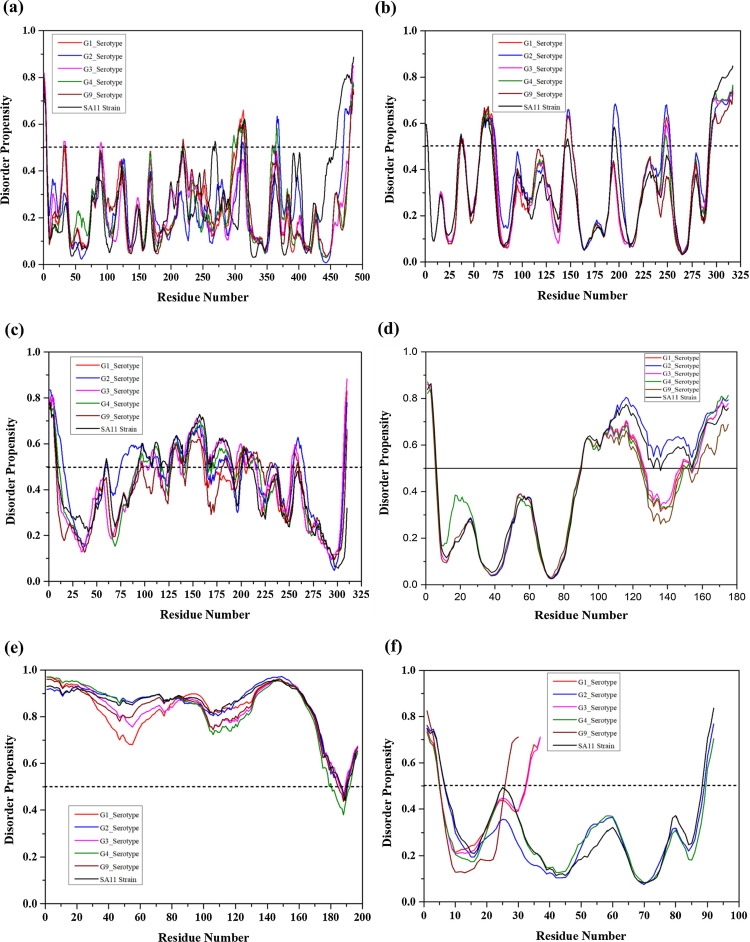
To evaluate if there are noticeable differences in the intrinsic disorder predisposition of rotaviral proteins from different serotypes of this virus, we conducted a computational analysis of structural and non-structural proteins from the retrovirus serotypes G1, G2, G3, G4, and G9. This comparative analysis supported the occurrence of a higher percentage of disordered regions in non-structural proteins and the presence of short IDPRs in structural proteins {Fig. 6, Fig. 7, Fig. 8 }. Although viral proteins from these serotypes have shown similar intrinsic disordered patterns as those examined in the SA11 reference strain, there also were some important differences. For example, the G2 serotype was characterized by slightly higher protein disorder percentage among all five serotypes, with the largest difference being observed for NSP2, NSP3, NSP4, NSP5, VP1, and VP6 of SA11 strain (see {Fig. 2.1, Fig. 2.2, Fig. 4.1, Fig. 4.2}).
Fig. 6.
Outcomes of the evaluation of the intrinsic disorder predisposition of rotavirus proteins from different serotypes in terms of the PONDR® VSL2-based PPID values. PPID scores for all proteins of rotavirus are specified on the y-axis. Results for proteins from G1, G2, G3, G4, and G5 serotypes are shown by red, blue, magenta, olive, and wine color bars, respectively. Corresponding results for the SA11 reference strain are shown by black bars.
Fig. 7.
Comparative analysis of the intrinsically disorder predisposition of the non-structural rotaviral proteins from 5 different serotypes of human rotavirus (G1, G2, G3, G4, and G9) with reference strain SA11. PONDR® VSL2 predictor was utilized in this study. The PONDR® VSL2-based disorder profiles of non-structural proteins from each serotype (G1, G2, G3, G4, and G9) are represented by red, blue, magenta, olive, and wine colors, respectively. Black curves show corresponding results for the proteins from the SA11 reference strain. Plots represent corresponding per-residue disorder propensities for (a) NSP1, (b) NSP2, (c) NSP3 (d) NSP4, (e) NSP5, and (f) NSP6. The NSP6 protein found to be truncated in G1, G3 and G9 serotypes having residue length less than 40 amino acids.
Fig. 8.
Comparative analysis of the intrinsically disorder predisposition of the structural rotaviral proteins from 5 different serotypes of human rotavirus (G1, G2, G3, G4, and G9) with reference strain SA11. PONDR® VSL2 predictor was utilized in this study. The PONDR® VSL2-based disorder profiles of non-structural proteins from each serotype (G1, G2, G3, G4, and G9) are represented by red, blue, magenta, olive, and wine colors, respectively. Black curves show corresponding results for the proteins from the SA11 reference strain. Plots represent the corresponding per-residue disorder propensities for (a) VP1/RdRp, (b) VP2, (c) VP3, (d) VP4, (e) VP6, and (f) VP7.
4. Conclusion
The functional mechanisms of some of the rotaviral proteins are known, but not completely deciphered because of the non-availability of complete structural information. The main objective of this analysis is to fill the gap to some extent by providing the data on the prevalence of intrinsic disorder in the form of intrinsically disordered proteins/regions (IDPs/IDPRs) in the rotavirus proteome. Based on several computational and experimental studies, it is evident that IDPRs or IDPs and MoRFs are crucial for viral replication and pathogenesis. Intrinsic disorder is also known to confer flexible conformations to viral proteins, as well as provide important mechanistic means for their binding promiscuity; i.e., their ability to be engaged in interactions with many viral and host proteins, as demonstrated for rotaviral NSP2 and NSP5 within the viroplasm [91], [133] and for proteins of many other viruses [141], leading to the subversion/hijacking of host cellular processes, including the protein synthesis machinery and nuclear transport to promote virus propagation [142], [143]. To compensate for the lack of the detailed information on the dark side of the rotaviral proteome, by employing several computational tools, here we show that rotavirus (the reference strain (SA11) and all five serotypes (G1, G2, G3, G4, and G9) analyzed in this study) contains several short and a few long IDPR in many structural and non-structural proteins, with NSP5 being expected to be disordered as a whole. Comparatively to other rotaviral proteins, NSP4 and NSP5 showed highest levels of intrinsic disorder that appear to be crucial for their multi-faceted functions. Also, the presence of multiple MoRFs predominantly in NSP5 indicates the functional importance of this protein for the rotavirus life cycle. Therefore, this study suggests that by virtue of its presence, intrinsic disorder (in the form of IDPRs with various length, or in the form of total disorder of NSP5) provides significant conformational plasticity and functional diversity to the rotaviral proteins and contributes to the assorted biological functions involving communication with various host proteins and protein-protein interactions within the virus. A well-known functional mechanism adopted by disordered proteins is disorder-to-order transitions upon binding to a partner that could be disrupted by small molecules. A new approach for drug design based on the concept of ligand-mediated disorder-to-order transition is slowly growing traction over traditional structure-based drug screening studies. Recent studies on the identification of small molecules that inhibit viroplasm formation by targeting protein-protein interactions [144] in this supramolecular rotavirus replication organelle could form the basis for the drug discovery based on the disordered regions of NSP2 and NSP5 that are essential for nucleation of viroplasm and recruitment of a large number of viral and host proteins into the viroplasm [91], [133]. Since high mutation rate among RNA viruses is the leading cause of vaccine failure, targeting disordered regions with small molecules for inhibiting either binding to their partners or preventing their conformational transition may provide a potential alternate therapeutic approach. We believe that understanding the penetrance of the IDPs/IDPRs in in rotavirus proteome would provide a new direction to decipher the mechanisms of virus pathogenesis, morphogenesis, virus survival, and drug discovery.
Acknowledgments
Acknowledgment
This work was partially supported by DST grant, India (YSS/2015/000613) to RG and IIT-Mandi, India to RG. DK is grateful to ICMR fellowship.
Author's contributions
RG, conception, design and study supervision; RG, VU, CDR, AS, PK and DK.: acquisition, analysis, and interpretation of data, writing, and review of the manuscript. # DK and AS contributed equally.
Declartation of Competing Interest
The authors declare no competing financial interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2019.09.166.
Contributor Information
C. Durga Rao, Email: cdrmcb@gmail.com.
Rajanish Giri, Email: rajanishgiri@iitmandi.ac.in.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Troeger C., Forouzanfar M., Rao P.C., Khalil I., Brown A., Reiner R.C., Fullman N., Thompson R.L., Abajobir A., Ahmed M., Alemayohu M.A., Alvis-Guzman N., Amare A.T., Antonio C.A., Asayesh H., Avokpaho E., Awasthi A., Bacha U., Barac A., Betsue B.D., Beyene A.S., Boneya D.J., Malta D.C., Dandona L., Dandona R., Dubey M., Eshrati B., Fitchett J.R.A., Gebrehiwot T.T., Hailu G.B., Horino M., Hotez P.J., Jibat T., Jonas J.B., Kasaeian A., Kissoon N., Kotloff K., Koyanagi A., Kumar G.A., Rai R.K., Lal A., El Razek H.M.A., Mengistie M.A., Moe C., Patton G., Platts-Mills J.A., Qorbani M., Ram U., Roba H.S., Sanabria J., Sartorius B., Sawhney M., Shigematsu M., Sreeramareddy C., Swaminathan S., Tedla B.A., Jagiellonian R.T.-M., Ukwaja K., Werdecker A., Widdowson M.-A., Yonemoto N., El Sayed Zaki M., Lim S.S., Naghavi M., Vos T., Hay S.I., Murray C.J.L., Mokdad A.H. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the global burden of disease study 2015. Lancet Infect. Dis. 2017;17:909–948. doi: 10.1016/S1473-3099(17)30276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO | Rotavirus, WHO, 2018.
- 3.Clark H.F., Lawley D.A., Schaffer A., Patacsil J.M., Marcello A.E., Glass R.I., Jain V., Gentsch J. Assessment of the epidemic potential of a new strain of rotavirus associated with the novel G9 serotype which caused an outbreak in the United States for the first time in the 1995–1996 season. J. Clin. Microbiol. 2004;42:1434–1438. doi: 10.1128/JCM.42.4.1434-1438.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Ryan M. The ever-changing landscape of rotavirus serotypes pediatr. Infect. Dis. J. 2009;28:S60–S62. doi: 10.1097/INF.0b013e3181967c29. [DOI] [PubMed] [Google Scholar]
- 5.Estes M.K., Cohen J. Rotavirus gene structure and function. Microbiol. Mol. Biol. Rev. 1989;53:410–449. doi: 10.1128/mr.53.4.410-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Settembre E.C., Chen J.Z., Dormitzer P.R., Grigorieff N., Harrison S.C. Atomic model of an infectious rotavirus particle. EMBO J. 2010;30:408–416. doi: 10.1038/emboj.2010.322. 10.1038/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad B.V., Wang G.J., Clerx J.P., Chiu W. Three-dimensional structure of rotavirus. J. Mol. Biol. 1988;199:269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- 8.Uversky V.N., Dunker A.K. Understanding protein non-folding. Biochim. Biophys. Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhowmick A., Brookes D.H., Yost S.R., Dyson H.J., Forman-Kay J.D., Gunter D., Head-Gordon M., Hura G.L., Pande V.S., Wemmer D.E., Wright P.E., Head-Gordon T. Finding our way in the dark proteome. J. Am. Chem. Soc. 2016;138:9730–9742. doi: 10.1021/jacs.6b06543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perdigão N., Heinrich J., Stolte C., Sabir K.S., Buckley M.J., Tabor B., Signal B., Gloss B.S., Hammang C.J., Rost B., Schafferhans A., O’Donoghue S.I. Unexpected features of the dark proteome. Proc. Natl. Acad. Sci. 2015;112:15898–15903. doi: 10.1073/pnas.1508380112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uversky V.N. Intrinsically disordered proteins from A to Z. Int. J. Biochem. Cell Biol. 2011;43:1090–1103. doi: 10.1016/j.biocel.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Xue B., Blocquel D., Habchi J., Uversky A.V., Kurgan L., Uversky V.N., Longhi S. Structural disorder in viral proteins. Chem. Rev. 2014;114:6880–6911. doi: 10.1021/cr4005692. [DOI] [PubMed] [Google Scholar]
- 13.Giri R., Kumar D., Sharma N., Uversky V.N. Intrinsically disordered side of the zika virus proteome. Front. Cell. Infect. Microbiol. 2016;6:144. doi: 10.3389/fcimb.2016.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh A., Kumar A., Yadav R., Uversky V.N., Giri R. Deciphering the dark proteome of Chikungunya virus. Sci. Rep. 2018;8:5822. doi: 10.1038/s41598-018-23969-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan X., Xue B., Dolan P.T., LaCount D.J., Kurgan L., Uversky V.N. 23713779. Mol. BioSyst. 2014;10:1345–1363. doi: 10.1039/c4mb00027g. [DOI] [PubMed] [Google Scholar]
- 16.Xue B., Mizianty M.J., Kurgan L., Uversky V.N. Protein intrinsic disorder as a flexible armor and a weapon of HIV-1. Cell. Mol. Life Sci. 2012;69:1211–1259. doi: 10.1007/s00018-011-0859-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue B., Ganti K., Rabionet A., Banks L., Uversky V.N. Disordered interactome of human papillomavirus. Curr. Pharm. Des. 2014;20:1274–1292. doi: 10.2174/13816128113199990072. [DOI] [PubMed] [Google Scholar]
- 18.Whelan J.N., Reddy K.D., Uversky V.N., Teng M.N. Functional correlations of respiratory syncytial virus proteins to intrinsic disorder. Mol. Biosyst. 2016;12:1507–1526. doi: 10.1039/c6mb00122j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng F., Badierah R.A., Almehdar H.A., Redwan E.M., Kurgan L., Uversky V.N. Unstructural biology of the Dengue virus proteins. FEBS J. 2015;282:3368–3394. doi: 10.1111/febs.13349. [DOI] [PubMed] [Google Scholar]
- 20.Goh G.K.-M., Dunker A.K., Uversky V. Prediction of intrinsic disorder in MERS-CoV/HCoV-EMC supports a high oral-fecal transmission. PLoS Curr. 2013 doi: 10.1371/currents.outbreaks.22254b58675cdebc256dbe3c5aa6498b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Redwan E.M., AlJaddawi A.A., Uversky V.N. Structural disorder in the proteome and interactome of Alkhurma virus (ALKV) Cell. Mol. Life Sci. 2018 doi: 10.1007/s00018-018-2968-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Babu M.M. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem. Soc. Trans. 2016;44:1185–1200. doi: 10.1042/BST20160172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oldfield C.J., Meng J., Yang J.Y., Yang M.Q., Uversky V.N., Dunker A.K. Flexible nets: disorder and induced fit in the associations of p53 and 14–3-3 with their partners. BMC Genomics. 2008;9:S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toto A., Camilloni C., Giri R., Brunori M., Vendruscolo M., Gianni S. Molecular recognition by templated folding of an intrinsically disordered protein. Sci. Rep. 2016;6:21994. doi: 10.1038/srep21994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar D., Sharma N., Giri R. Therapeutic interventions of cancers using intrinsically disordered proteins as drug targets: C-myc as model system. Cancer Inform. 2017;16 doi: 10.1177/1176935117699408. 1176935117699408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunori M., Gianni S., Giri R., Morrone A., Travaglini-Allocatelli C. Morphogenesis of a protein: folding pathways and the energy landscape. Biochem. Soc. Trans. 2012;40:429–432. doi: 10.1042/BST20110683. [DOI] [PubMed] [Google Scholar]
- 27.Giri R., Morrone A., Toto A., Brunori M., Gianni S. Structure of the transition state for the binding of c-Myb and KIX highlights an unexpected order for a disordered system. Proc. Natl. Acad. Sci. U. S. A. 2013;110:14942–14947. doi: 10.1073/pnas.1307337110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oldfield C.J., Cheng Y., Cortese M.S., Romero P., Uversky V.N., Dunker A.K. Coupled folding and binding with α-helix-forming molecular recognition elements †. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 29.Mohan A., Oldfield C.J., Radivojac P., Vacic V., Cortese M.S., Dunker A.K., Uversky V.N. Analysis of molecular recognition features (MoRFs) J. Mol. Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 30.Cheng Y., Oldfield C.J., Meng J., Romero P., Uversky V.N., Dunker A.K. Mining?-helix-forming molecular recognition features with cross species sequence alignments? Biochemistry. 2007;46:13468–13477. doi: 10.1021/bi7012273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra P.M., Uversky V.N., Giri R. Molecular recognition features in zika virus proteome. J. Mol. Biol. 2017 doi: 10.1016/j.jmb.2017.10.018. [DOI] [PubMed] [Google Scholar]
- 32.Singh A., Kumar A., Uversky V.N., Giri R. Understanding the interactability of chikungunya virus proteins via molecular recognition feature analysis. RSC Adv. 2018;8:27293–27303. doi: 10.1039/c8ra04760j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aarthy M., Kumar D., Giri R., Singh S.K. E7 oncoprotein of human papillomavirus: Structural dynamics and inhibitor screening study. Gene. 2018;658:159–177. doi: 10.1016/j.gene.2018.03.026. https://www.sciencedirect.com/science/article/pii/S0378111918302646#f0030 (accessed March 23, 2018) [DOI] [PubMed] [Google Scholar]
- 34.Sharma N., Murali A., Singh S.K., Giri R. Epigallocatechin gallate, an active green tea compound inhibits the Zika virus entry into host cells via binding the envelope protein. Int. J. Biol. Macromol. 2017;104:1046–1054. doi: 10.1016/j.ijbiomac.2017.06.105. [DOI] [PubMed] [Google Scholar]
- 35.López S., Arias C.F. Simian rotavirus SA11 strains. J. Virol. 1992;66:1832. doi: 10.1128/jvi.66.3.1832-.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hewish M., Takada Y., Coulson B. Integrins a2b1 and a4b1 can mediate SA11 rotavirus attachment and entry into cells. J. Virol. 2000 doi: 10.1128/jvi.74.1.228-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isa P., Arias C.F., López S. Role of sialic acids in rotavirus infection. Glycoconj. J. 2006 doi: 10.1007/s10719-006-5435-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dosztanyi Z., Csizmok V., Tompa P., Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 39.Dosztányi Z., Mészáros B., Simon I. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009;25:2745–2746. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Disfani F.M., Hsu W.-L., Mizianty M.J., Oldfield C.J., Xue B., Dunker A.K., Uversky V.N., Kurgan L. MoRFpred, a computational tool for sequence-based prediction and characterization of short disorder-to-order transitioning binding regions in proteins. Bioinformatics. 2012;28:i75–i83. doi: 10.1093/bioinformatics/bts209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malhis N., Jacobson M., Gsponer J. MoRFchibi SYSTEM: software tools for the identification of MoRFs in protein sequences. Nucleic Acids Res. 2016;44:W488–W493. doi: 10.1093/nar/gkw409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Linding R., Jensen L.J., Diella F., Bork P., Gibson T.J., Russell R.B. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. http://www.ncbi.nlm.nih.gov/pubmed/14604535 (accessed January 17, 2017) [DOI] [PubMed] [Google Scholar]
- 43.Linding R., Russell R.B., Neduva V., Gibson T.J. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward J.J., Sodhi J.S., McGuffin L.J., Buxton B.F., Jones D.T. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Romero P., Obradovic Z., Li X., Garner E.C., Brown C.J., Dunker A.K. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. http://www.ncbi.nlm.nih.gov/pubmed/11093259 (accessed January 17, 2017) [DOI] [PubMed] [Google Scholar]
- 46.Obradovic Z., Peng K., Vucetic S., Radivojac P., Dunker A.K. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins Struct. Funct. Bioinforma. 2005;61:176–182. doi: 10.1002/prot.20735. [DOI] [PubMed] [Google Scholar]
- 47.Peng K., Vucetic S., Radivojac P., Brown C.J., Dunker A.K., Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 2005;3:35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- 48.Xue B., Dunbrack R.L., Williams R.W., Dunker A.K., Uversky V.N. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferron F., Longhi S., Canard B., Karlin D., Kim S., Kanehisa M., Cortese M., Lawson J., Brown C., Sikes J.G., Newton C., Dunker A. A practical overview of protein disorder prediction methods. Proteins Struct. Funct. Bioinforma. 2006;65:1–14. doi: 10.1002/prot.21075. [DOI] [PubMed] [Google Scholar]
- 50.Deng X., Eickholt J., Cheng J. A comprehensive overview of computational protein disorder prediction methods. Mol. Biosyst. 2012;8:114–121. doi: 10.1039/c1mb05207a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.E. Gasteiger, C. Hoogland, A. Gattiker, S. Duvaud, M.R. Wilkins, R.D. Appel, A. Bairoch, Protein Identification and Analysis Tools on the ExPASy Server, in: Proteomics Protoc. Handb., 2009. doi:10.1385/1-59259-890-0:571.
- 52.Peng K., Radivojac P., Vucetic S., Dunker A.K., Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinf. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vacic V., Oldfield C.J., Mohan A., Radivojac P., Cortese M.S., Uversky V.N., Dunker A.K. Characterization of molecular recognition features, MoRFs, and their binding partners. J. Proteome Res. 2007;6:2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ward J.J., McGuffin L.J., Bryson K., Buxton B.F., Jones D.T. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004 doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 55.Malhis N., Wong E.T.C., Nassar R., Gsponer J. Computational identification of MoRFs in protein sequences using Hierarchical application of bayes rule. PLoS ONE. 2015 doi: 10.1371/journal.pone.0141603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011 doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dinkel H., Van Roey K., Michael S., Davey N.E., Weatheritt R.J., Born D., Speck T., Krüger D., Grebnev G., Kubań M., Strumillo M., Uyar B., Budd A., Altenberg B., Seiler M., Chemes L.B., Glavina J., Sánchez I.E., Diella F., Gibson T.J. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 2014 doi: 10.1093/nar/gkt1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dinkel H., Michael S., Weatheritt R.J., Davey N.E., Van Roey K., Altenberg B., Toedt G., Uyar B., Seiler M., Budd A., Jödicke L., Dammert M.A., Schroeter C., Hammer M., Schmidt T., Jehl P., McGuigan C., Dymecka M., Chica C., Luck K., Via A., Chatr-Aryamontri A., Haslam N., Grebnev G., Edwards R.J., Steinmetz M.O., Meiselbach H., Diella F., Gibson T.J. ELM–the database of eukaryotic linear motifs. Nucleic Acids Res. 2012;40:D242–D251. doi: 10.1093/nar/gkr1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu X., McDonald S.M., Tortorici M.A., Tao Y.J., Vasquez-Del Carpio R., Nibert M.L., Patton J.T., Harrison S.C. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure. 2008:1678–1688. doi: 10.1016/j.str.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McKell A.O., LaConte L.E.W., McDonald S.M. A temperature-sensitive lesion in the N-terminal domain of the rotavirus polymerase affects its intracellular localization and enzymatic activity. J. Virol. 2017;91 doi: 10.1128/JVI.00062-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jayaram H., Estes M., Prasad B.V.V. Emerging themes in rotavirus cell entry, genome organization, transcription and replication. Virus Res. 2004;101:67–81. doi: 10.1016/j.virusres.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 62.Patton J.T., Jones M.T., Kalbach A.N., He Y.W., Xiaobo J. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J. Virol. 1997;71:9618–9626. doi: 10.1128/jvi.71.12.9618-9626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McDonald S.M., Patton J.T. Rotavirus VP2 core shell regions critical for viral polymerase activation. J. Virol. 2011;85:3095–3105. doi: 10.1128/JVI.02360-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.J.A. Lawton, C.Q.-y. Zeng, S.K. Mukherjee, J. Cohen, M.K. Estes, B.V. Venkataram Prasad, Three-dimensional structural analysis of recombinant rotavirus-like particles with intact and amino-terminal-deleted VP2: Implications for the architecture of the VP2 capsid layer downloaded from, 1997. [DOI] [PMC free article] [PubMed]
- 65.Liu M., Offit P.A., Estes M.K. Identification of the simian rotavirus SA11 genome segment 3 product. Virology. 1988;163:26–32. doi: 10.1016/0042-6822(88)90230-9. [DOI] [PubMed] [Google Scholar]
- 66.Ogden K.M., Snyder M.J., Dennis A.F., Patton J.T. Predicted structure and domain organization of rotavirus capping enzyme and innate immune antagonist VP3. J. Virol. 2014;88:9072–9085. doi: 10.1128/JVI.00923-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brandmann T., Jinek M. Crystal structure of the C-terminal 2′,5′-phosphodiesterase domain of group a rotavirus protein VP3. Proteins Struct. Funct. Bioinforma. 2015;83:997–1002. doi: 10.1002/prot.24794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang W., Donnelly B., Bondoc A., Mohanty S.K., McNeal M., Ward R., Sestak K., Zheng S., Tiao G. The rhesus rotavirus gene encoding VP4 Is a major determinant in the pathogenesis of biliary atresia in newborn mice. J. Virol. 2011;85:9069–9077. doi: 10.1128/JVI.02436-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kindler E., Trojnar E., Heckel G., Otto P.H., Johne R. Analysis of rotavirus species diversity and evolution including the newly determined full-length genome sequences of rotavirus F and G. Infect. Genet. Evol. 2013;14:58–67. doi: 10.1016/j.meegid.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 70.Crawford S.E., Mukherjee S.K., Estes M.K., Lawton J.A., Shaw A.L., Ramig R.F., Prasad B.V. Trypsin cleavage stabilizes the rotavirus VP4 spike. J. Virol. 2001;75:6052–6061. doi: 10.1128/JVI.75.13.6052-6061.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lopez S., Arias C.F., Bell J.R., Strauss J.H., Espejo R.T. Primary structure of the cleavage site associated with trypsin enhancement of rotavirus SA11 infectivity. Virology. 1985;144:11–19. doi: 10.1016/0042-6822(85)90300-9. [DOI] [PubMed] [Google Scholar]
- 72.Estes M.K., Graham D.Y., Mason B.B. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J. Virol. 1981;39:879–888. doi: 10.1128/jvi.39.3.879-888.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dormitzer P.R., Nason E.B., Venkataram Prasad B.V., Harrison S.C. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature. 2004;430:1053–1058. doi: 10.1038/nature02836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dowling W., Denisova E., LaMonica R., Mackow E.R. Selective membrane permeabilization by the rotavirus VP5* protein is abrogated by mutations in an internal hydrophobic domain. J. Virol. 2000;74:6368–6376. doi: 10.1128/jvi.74.14.6368-6376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hoshino Y., Saif L.J., Kang S.-Y., Sereno M.M., Chen W.-K., Kapikian A.Z. Identification of Group A rotavirus genes associated with virulence of a porcine rotavirus and host range restriction of a human rotavirus in the gnotobiotic piglet model. Virology. 1995;209:274–280. doi: 10.1006/viro.1995.1255. [DOI] [PubMed] [Google Scholar]
- 76.Morelli M., Ogden K.M., Patton J.T. Silencing the alarms: Innate immune antagonism by rotavirus NSP1 and VP3. Virology. 2015;479–480:75–84. doi: 10.1016/j.virol.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fiore L., Greenberg H.B., Mackow E.R. The VP8 fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology. 1991;181:553–563. doi: 10.1016/0042-6822(91)90888-i. [DOI] [PubMed] [Google Scholar]
- 78.Nejmeddine M., Trugnan G., Sapin C., Kohli E., Svensson L., Lopez S., Cohen J. Rotavirus spike protein VP4 is present at the plasma membrane and is associated with microtubules in infected cells. J. Virol. 2000;74:3313–3320. doi: 10.1128/jvi.74.7.3313-3320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Condemine W., Eguether T., Couroussé N., Etchebest C., Gardet A., Trugnan G., Chwetzoff S. The C-terminus of rotavirus VP4 protein contains an actin binding domain which requires co-operation with the coiled-coil domain for actin remodeling. J. Virol. 2018 doi: 10.1128/JVI.01598-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Greenberg H., McAuliffe V., Valdesuso J., Wyatt R., Flores J., Kalica A., Hoshino Y., Singh N. Serological analysis of the subgroup protein of rotavirus, using monoclonal antibodies. Infect. Immun. 1983;39:91–99. doi: 10.1128/iai.39.1.91-99.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clapp L.L., Patton J.T. Rotavirus morphogenesis: domains in the major inner capsid protein essential for binding to single-shelled particles and for trimerization. Virology. 1991;180:697–708. doi: 10.1016/0042-6822(91)90083-n. [DOI] [PubMed] [Google Scholar]
- 82.Ginn D.I., Ward R.L., Hamparian V.V., Hughes J.H. Inhibition of rotavirus in vitro transcription by optimal concentrations of monoclonal antibodies specific for rotavirus VP6. J. Gen. Virol. 1992;73:3017–3022. doi: 10.1099/0022-1317-73-11-3017. [DOI] [PubMed] [Google Scholar]
- 83.J.Z. Chen, E.C. Settembre, S.T. Aoki, X. Zhang, A.R. Bellamy, P.R. Dormitzer, S.C. Harrison, N. Grigorieff, Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM, (n.d.). [DOI] [PMC free article] [PubMed]
- 84.Trask S.D., Dormitzer P.R. Assembly of highly infectious rotavirus particles recoated with recombinant outer capsid proteins. J. Virol. 2006;80:11293–11304. doi: 10.1128/JVI.01346-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mathieu M., Petitpas I., Navaza J., Lepault J., Kohli E., Pothier P., Prasad B.V.V., Cohen J., Rey F.A. Atomic structure of the major capsid protein of rotavirus: implications for the architecture of the viron. EMBO J. 2001;20:1485–1497. doi: 10.1093/emboj/20.7.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hua J., Mansell E.A., Patton J.T. Comparative analysis of the rotavirus NS53 gene: Conservation of basic and cysteine-rich regions in the protein and possible stem-loop structures in the RNA. Virology. 1993;196:372–378. doi: 10.1006/viro.1993.1492. [DOI] [PubMed] [Google Scholar]
- 87.Hua J., Chen X., Patton J.T. Deletion mapping of the rotavirus metalloprotein NS53 (NSP1): the conserved cysteine-rich region is essential for virus-specific RNA binding. J. Virol. 1994;68:3990–4000. doi: 10.1128/jvi.68.6.3990-4000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Taniguchi K., Kojima K., Kobayashi N., Urasawa T., Urasawa S. Viral Gastroenteritis. Springer Vienna; Vienna: 1996. pp. 53–58. [Google Scholar]
- 89.Aponte C., Poncet D., Cohen J. Recovery and characterization of a replicase complex in rotavirus-infected cells by using a monoclonal antibody against NSP2 downloaded from. J. Virol. 1996;70:985–991. doi: 10.1128/jvi.70.2.985-991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eichwald C., Rodriguez J.F., Burrone O.R. Characterization of rotavirus NSP2/NSP5 interactions and the dynamics of viroplasm formation. J. Gen. Virol. 2004;85:625–634. doi: 10.1099/vir.0.19611-0. [DOI] [PubMed] [Google Scholar]
- 91.Dhillon P., Tandra V.N., Chorghade S.G., Namsa N.D., Sahoo L., Rao C.D. Cytoplasmic relocalization and colocalization with viroplasms of host cell proteins, and their role in rotavirus infection. J. Virol. 2018;92 doi: 10.1128/JVI.00612-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Eichwald C., Arnoldi F., Laimbacher A.S., Schraner E.M., Fraefel C., Wild P., Burrone O.R., Ackermann M. Rotavirus viroplasm fusion and perinuclear localization are dynamic processes requiring stabilized microtubules. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0047947. e47947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Martin D., Duarte M., Lepault J., Poncet D. Sequestration of free tubulin molecules by the viral protein NSP2 induces microtubule depolymerization during rotavirus infection. J. Virol. 2010;84:2522–2532. doi: 10.1128/JVI.01883-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jayaram H., Taraporewala Z., Patton J.T., Prasad B.V.V. Rotavirus protein involved in genome replication and packaging exhibits a HIT-like fold. Nature. 2002;417:311–315. doi: 10.1038/417311a. [DOI] [PubMed] [Google Scholar]
- 95.Arnoldi F., Campagna M., Eichwald C., Desselberger U., Burrone O.R. Interaction of rotavirus polymerase VP1 with nonstructural protein NSP5 is stronger than that with NSP2. J. Virol. 2007;81:2128–2137. doi: 10.1128/JVI.01494-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taraporewala Z., Chen D., Patton J.T. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J. Virol. 1999;73:9934–9943. doi: 10.1128/jvi.73.12.9934-9943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Taraporewala Z.F., Jiang X., Vasquez-Del Carpio R., Jayaram H., Prasad B.V.V., Patton J.T. Structure-function analysis of rotavirus NSP2 octamer by using a novel complementation system. J. Virol. 2006;80:7984–7994. doi: 10.1128/JVI.00172-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taraporewala Z.F., Patton J.T. Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2. J. Virol. 2001;75:4519–4527. doi: 10.1128/JVI.75.10.4519-4527.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rao C., Das M., Ilango P., Lalwani R., Rao B., Gowda K. Comparative nucleotide and amino acid sequence analysis of the sequence-specific RNA-binding rotavirus nonstructural protein NSP3. Virology. 1995;207:327–333. doi: 10.1006/viro.1995.1087. [DOI] [PubMed] [Google Scholar]
- 100.Poncet D., Aponte C., Cohen J. Rotavirus protein NSP3 (NS34) is bound to the 3’ end consensus sequence of viral mRNAs in infected cells. J. Virol. 1993;67:3159–3165. doi: 10.1128/jvi.67.6.3159-3165.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mattion N.M., Cohen J., Aponte C., Estes M.K. Characterization of an oligomerization domain and RNA-binding properties on rotavirus nonstructural protein NS34. Virology. 1992;190:68–83. doi: 10.1016/0042-6822(92)91193-x. [DOI] [PubMed] [Google Scholar]
- 102.Piron M., Delaunay T., Grosclaude J., Poncet D. Identification of the RNA-binding, dimerization, and eIF4GI-binding domains of rotavirus nonstructural protein NSP3. J. Virol. 1999;73:5411–5421. doi: 10.1128/jvi.73.7.5411-5421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Deo R.C., Groft C.M., Rajashankar K.R., Burley S.K. Recognition of the rotavirus mRNA 3′ consensus by an asymmetric NSP3 homodimer. Cell. 2002;108:71–81. doi: 10.1016/s0092-8674(01)00632-8. [DOI] [PubMed] [Google Scholar]
- 104.Groft C.M., Burley S.K. Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol. Cell. 2002;9:1273–1283. doi: 10.1016/s1097-2765(02)00555-5. [DOI] [PubMed] [Google Scholar]
- 105.Padilla-Noriega L., Paniagua O., Guzmán-León S. Rotavirus protein NSP3 shuts off host cell protein synthesis. Virology. 2002;298:1–7. doi: 10.1006/viro.2002.1477. [DOI] [PubMed] [Google Scholar]
- 106.Piron M., Vende P., Cohen J., Poncet D. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 1998;17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Contreras-Treviño H.I., Reyna-Rosas E., León-Rodríguez R., Ruiz-Ordaz B.H., Dinkova T.D., Cevallos A.M., Padilla-Noriega L. Species A rotavirus NSP3 acquires its translation inhibitory function prior to stable dimer formation. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0181871. e0181871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Au K.S., Chan W.K., Burns J.W., Estes M.K. Receptor activity of rotavirus nonstructural glycoprotein NS28. J. Virol. 1989;63:4553–4562. doi: 10.1128/jvi.63.11.4553-4562.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bugarcic A., Taylor J.A. Rotavirus nonstructural glycoprotein NSP4 is secreted from the apical surfaces of polarized epithelial cells. J. Virol. 2006;80:12343–12349. doi: 10.1128/JVI.01378-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ball J.M., Tian P., Zeng C.Q., Morris A.P., Estes M.K. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science. 1996;272:101–104. doi: 10.1126/science.272.5258.101. [DOI] [PubMed] [Google Scholar]
- 111.Jagannath M.R., Kesavulu M.M., Deepa R., Sastri P.N., Senthil Kumar S., Suguna K., Durga Rao C. N-and C-terminal cooperation in rotavirus enterotoxin: novel mechanism of modulation of the properties of a multifunctional protein by a structurally and functionally overlapping conformational domain. J. Virol. 2006;80:412–425. doi: 10.1128/JVI.80.1.412-425.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Durga Rao C., Pamidimukkala K., Marathahalli J.R., Kaza S., Rao C.D. Conformational differences unfold a wide range of enterotoxigenic abilities exhibited by rNSP4 peptides from different rotavirus strains. Open Virol. J. 2011;5:124–135. doi: 10.2174/1874357901105010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rajasekaran D., Sastri N.P., Marathahalli J.R., Indi S.S., Pamidimukkala K., Suguna K., Rao C.D. The flexible C terminus of the rotavirus non-structural protein NSP4 is an important determinant of its biological properties. J. Gen. Virol. 2008;89:1485–1496. doi: 10.1099/vir.0.83617-0. [DOI] [PubMed] [Google Scholar]
- 114.Yang W., McCrae M.A. The rotavirus enterotoxin (NSP4) promotes re-modeling of the intracellular microtubule network. Virus Res. 2012;163:269–274. doi: 10.1016/j.virusres.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 115.Xu A., Bellamy A.R., Taylor J.A. Immobilization of the early secretory pathway by a virus glycoprotein that binds to microtubules. EMBO J. 2000;19:6465–6474. doi: 10.1093/emboj/19.23.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Uversky V.N. A decade and a half of protein intrinsic disorder: biology still waits for physics. Protein Sci. 2013;22:693–724. doi: 10.1002/pro.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Huang H., Schroeder F., Estes M.K., Mcpherson T., Ball J.M. Interaction(s) of rotavirus non-structural protein 4 (NSP4) C-terminal peptides with model membranes. Biochem. J. 2004 doi: 10.1042/BJ20031789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chacko A.R., Arifullah M., Sastri N.P., Jeyakanthan J., Ueno G., Sekar K., Read R.J., Dodson E.J., Rao D.C., Suguna K. Novel pentameric structure of the diarrhea-inducing region of the rotavirus enterotoxigenic protein NSP4. J. Virol. 2011;85:12721–12732. doi: 10.1128/JVI.00349-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chacko A.R., Jeyakanthan J., Ueno G., Sekar K., Rao C.D., Dodson E.J., Suguna K., Read R.J. A new pentameric structure of rotavirus NSP4 revealed by molecular replacement. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012;68:57–61. doi: 10.1107/S0907444911049705. [DOI] [PubMed] [Google Scholar]
- 120.Deepa R., Durga Rao C., Suguna K. Structure of the extended diarrhea-inducing domain of rotavirus enterotoxigenic protein NSP4. Arch. Virol. 2007;152:847–859. doi: 10.1007/s00705-006-0921-x. [DOI] [PubMed] [Google Scholar]
- 121.Bowman G.D., Nodelman I.M., Levy O., Lin S.L., Tian P., Zamb T.J., Udem S.A., Venkataraghavan B., Schutt C.E. Crystal structure of the oligomerization domain of NSP4 from rotavirus reveals a core metal-binding site. J. Mol. Biol. 2000;304:861–871. doi: 10.1006/jmbi.2000.4250. [DOI] [PubMed] [Google Scholar]
- 122.Kumar S., Ramappa R., Pamidimukkala K., Rao C.D., Suguna K. New tetrameric forms of the rotavirus NSP4 with antiparallel helices. Arch. Virol. 2018;163:1531–1547. doi: 10.1007/s00705-018-3753-6. [DOI] [PubMed] [Google Scholar]
- 123.Parr R.D., Storey S.M., Mitchell D.M., McIntosh A.L., Zhou M., Mir K.D., Ball J.M. The rotavirus enterotoxin NSP4 directly interacts with the caveolar structural protein caveolin-1. J. Virol. 2006;80:2842–2854. doi: 10.1128/JVI.80.6.2842-2854.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Welch S.K., Crawford S.E., Estes M.K. Rotavirus SA11 genome segment 11 protein is a nonstructural phosphoprotein. J. Virol. 1989;63:3974–3982. doi: 10.1128/jvi.63.9.3974-3982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.González S.A., Burrone O.R. Rotavirus NS26 is modified by addition of single O-linked residues of N-acetylglucosamine. Virology. 1991;182:8–16. doi: 10.1016/0042-6822(91)90642-o. [DOI] [PubMed] [Google Scholar]
- 126.Afrikanova I., Miozzo M.C., Giambiagi S., Burrone O. Phosphorylation generates different forms of rotavirus NSP5. J. Gen. Virol. 1996;77:2059–2065. doi: 10.1099/0022-1317-77-9-2059. [DOI] [PubMed] [Google Scholar]
- 127.Eichwald C., Vascotto F., Fabbretti E., Burrone O.R. Rotavirus NSP5: mapping phosphorylation sites and kinase activation and viroplasm localization domains. J. Virol. 2002;76:3461–3470. doi: 10.1128/JVI.76.7.3461-3470.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Afrikanova I., Fabbretti E., Miozzo M.C., Burrone O.R. Printed in great britain rotavirus NSP5 phosphorylation is up-regulated by interaction with NSP2. J. Gen. Virol. 2018;79:2679–2686. doi: 10.1099/0022-1317-79-11-2679. [DOI] [PubMed] [Google Scholar]
- 129.Contin R., Arnoldi F., Campagna M., Burrone O.R. Rotavirus NSP5 orchestrates recruitment of viroplasmic proteins. J. Gen. Virol. 2010;91:1782–1793. doi: 10.1099/vir.0.019133-0. [DOI] [PubMed] [Google Scholar]
- 130.Mohan K.V.K., Muller J., Som I., Atreya C.D. The N- and C-terminal regions of rotavirus NSP5 are the critical determinants for the formation of viroplasm-like structures independent of NSP2. J. Virol. 2003;77:12184–12192. doi: 10.1128/JVI.77.22.12184-12192.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Blackhall J., Fuentes A., Hansen K., Magnusson G. Serine protein kinase activity associated with rotavirus. Phosphoprotein NSP5. 1997 doi: 10.1128/jvi.71.1.138-144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fuerst T.R., Niles E.G., Studier F.W., Moss B., Burrone O.R. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dhillon P., Durga Rao C. Rotavirus induces formation of remodeled stress granules and P-bodies and their sequestration in viroplasms to promote progeny virus production. J. Virol. 2018 doi: 10.1128/JVI.01363-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rancurel C., Khosravi M., Dunker A.K., Romero P.R., Karlin D. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J. Virol. 2009;83:10719–10736. doi: 10.1128/JVI.00595-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xue B., Williams R.W., Oldfield C.J., Goh G.K.-M., Dunker A.K., Uversky V.N. Viral disorder or disordered viruses: do viral proteins possess unique features? Protein Pept. Lett. 2010;17:932–951. doi: 10.2174/092986610791498984. [DOI] [PubMed] [Google Scholar]
- 136.Mitchell D.B., Both G.W. Simian rotavirus SA11 segment 11 contains overlapping reading frames. Nucleic Acids Res. 1988;16:6244. doi: 10.1093/nar/16.13.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Heiman E.M., McDonald S.M., Barro M., Taraporewala Z.F., Bar-Magen T., Patton J.T. Group A human rotavirus genomics: evidence that gene constellations are influenced by viral protein interactions. J. Virol. 2008 doi: 10.1128/JVI.01402-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rainsford E.W., McCrae M.A. Characterization of the NSP6 protein product of rotavirus gene 11. Virus Res. 2007;130:193–201. doi: 10.1016/j.virusres.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 139.G. Holloway, R.I. Johnson, Y. Kang, V.T. Dang, D. Stojanovski, B.S. Coulson, Rotavirus NSP6 localizes to mitochondria via a predicted N-terminal a-helix, 2018, doi:10.1099/jgv.0.000294. [DOI] [PubMed]
- 140.Komoto S., Kanai Y., Fukuda S., Kugita M., Kawagishi T., Ito N., Sugiyama M., Matsuura Y., Kobayashi T., Taniguchi K. Reverse genetics system demonstrates that rotavirus nonstructural protein NSP6 is not essential for viral replication in cell culture. J. Virol. 2017;91 doi: 10.1128/JVI.00695-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lloyd R.E. Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses. Virology. 2015;479–480:457–474. doi: 10.1016/j.virol.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Walsh D., Mohr I. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 2011;9:860–875. doi: 10.1038/nrmicro2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yarbrough M.L., Mata M.A., Sakthivel R., Fontoura B.M.A. Viral subversion of nucleocytoplasmic trafficking. Traffic. 2014;15:127–140. doi: 10.1111/tra.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.La Frazia S., Ciucci A., Arnoldi F., Coira M., Gianferretti P., Angelini M., Belardo G., Burrone O.R., Rossignol J.-F., Santoro M.G. Thiazolides, a new class of antiviral agents effective against rotavirus infection, target viral morphogenesis, inhibiting viroplasm formation. J. Virol. 2013;87:11096–11106. doi: 10.1128/JVI.01213-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.