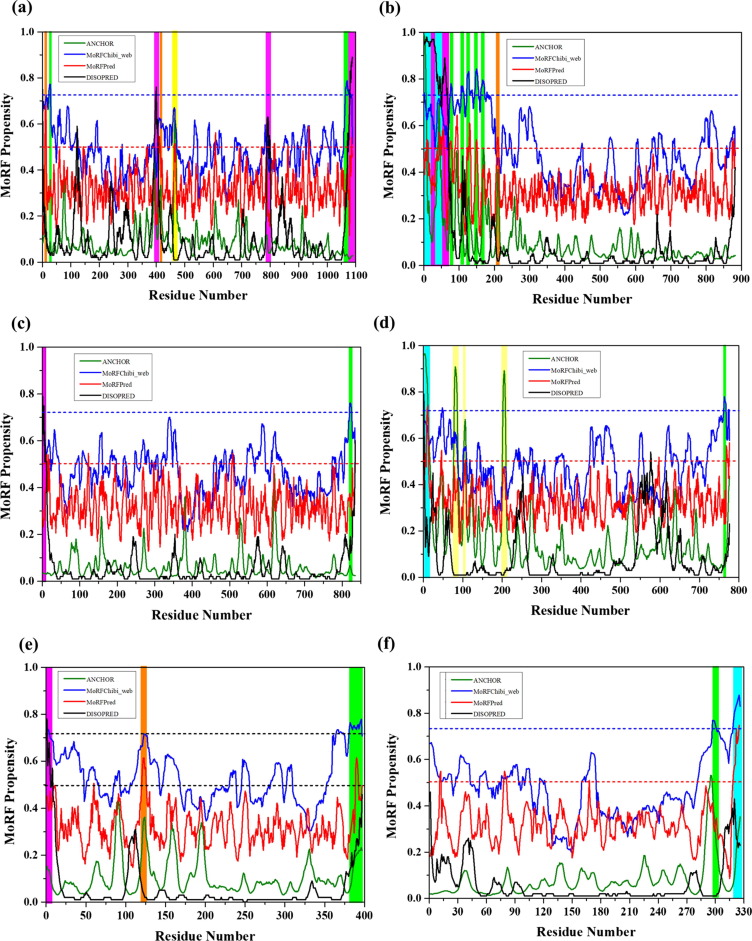
Fig. 3.
Evaluation of disorder-based interactivity of the rotavirus structural proteins. The presence of MoRF regions was evaluated by ANCHOR (olive lines), DISOPRED (black line), MoRFpred (red line) and MoRFchibi_web blue lines). The threshold for MoRF predictions by ANCHOR, DISOPRED, MoRFPred are 0.5 and of MoRFchibi_web is 0.725. These thresholds are shown as dashed red lines and blue line, respectively. Positions of MoRFs predicted by ANCHOR is shown by a yellow bar, orange bar by MoRFPred, pink bar by DISOPRED, overlapped common MoRF positions are shown by cyan bar, and MoRFchibi_web are shown by olive bars, respectively. Plots signify MoRF analysis of (a) VP1 (b) VP2 (c) VP3 (d) VP4 (e) VP6 (f) VP7.