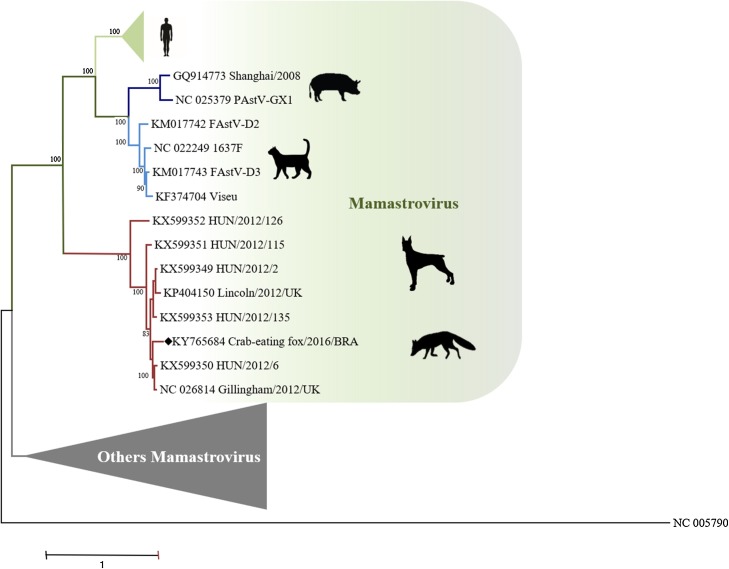
Fig. 3.
The whole genome phylogenetic tree.
Nucleotide phylogenetic tree (A) was reconstructed using General Time Reversible (GTR) model. Gamma distribution with invariant site (G + I) were applied to both inferences. The percentage of replicate trees in which the associated taxa are grouped in the bootstrap test (1000 replicates) is shown next to the branches. GenBank accession numbers are listed for all sequences analyzed in the tree. The crab-eating fox/2016/BRA sequence is labeled with a black diamond (♦).