Extended Data Fig. 6. Independent repeat of 13C carbon tracing by LC-HRMS and Hexokinase-2 mRNA expression in support of Fig. 5.
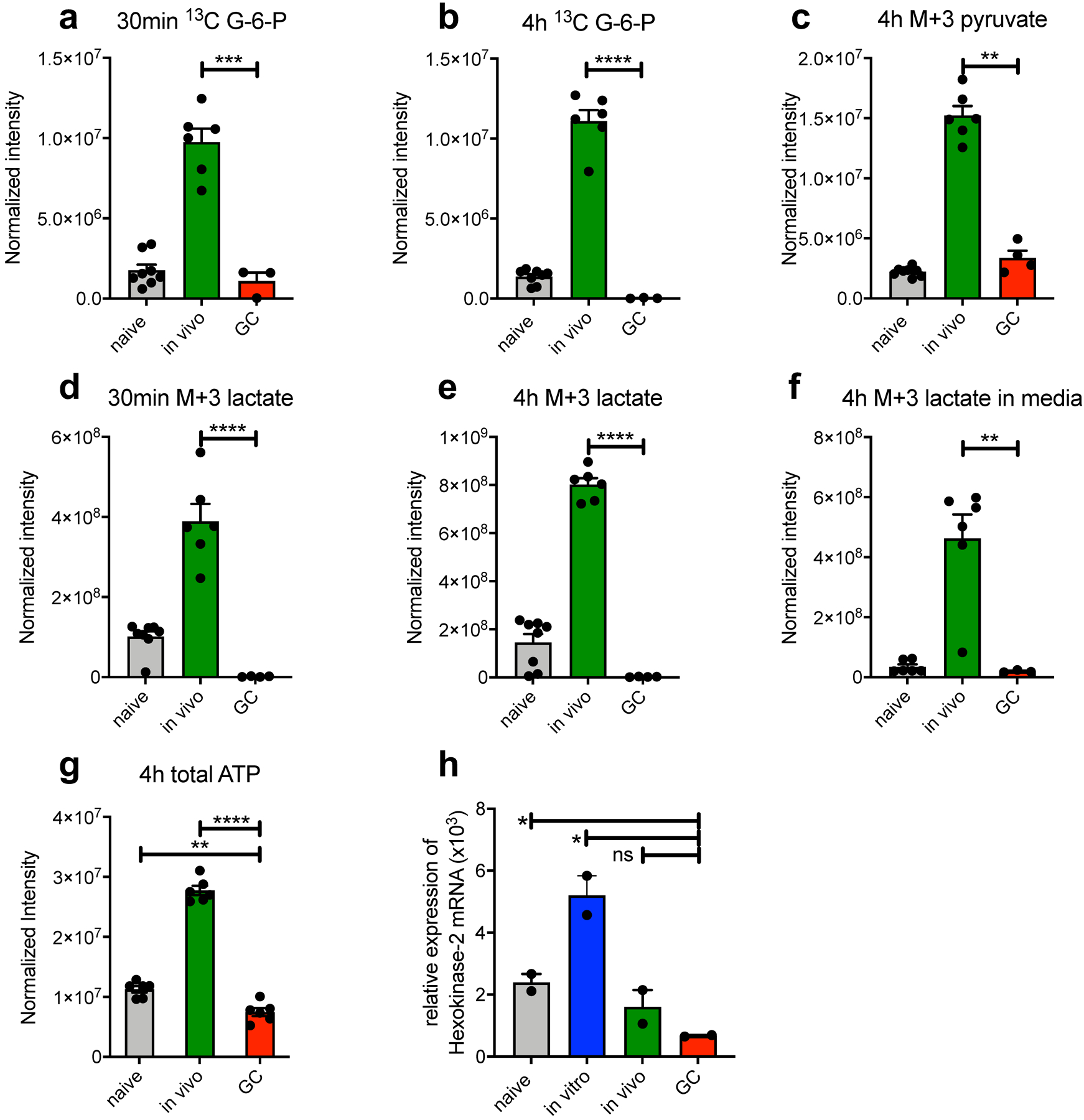
(a-g) Bead purified naive, in vivo activated and GC B cells were stimulated with anti-CD40 in glucose and glutamine free RPMI media with the addition of 2 mg/ml [13C6]-glucose for 30min or 4h. Cells (a-e, g) and supernatants (f) were then subjected to LC-HRMS. Depicted is one representative experiment with n=6 per sample. Each n represents a pool of 3 individual wells. (H) qRT-PCR for Hexokinase-2 expression from freshly isolated cell populations relative to RPS9. Bars are means +/−SEM. Normalization was performed as described in the methods section; ns = not significant; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001 by unpaired, two-tailed t-test.
