Fig. 5. Phylogenomic analysis of prasinovirus OmV2.
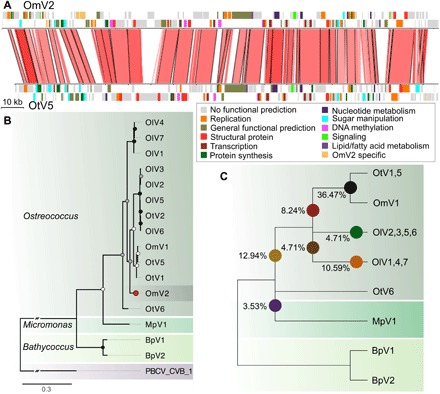
(A) Whole-genome alignment of OmV2 compared to the O. tauri virus, OtV5. Genomic regions with 60 to 100% nucleotide identity are joined by red blocks with higher identity indicated by darker red. Genes are colored according to functional groups. (B) Maximum likelihood phylogenetic tree of amino acid sequences of the DNA polymerase B gene from prasinoviruses infecting the genera Ostreococcus, Micromonas, and Bathycoccus. The tree was rooted with PBCV-1 (Chlorovirus) with the connecting branch truncated for display. The red circle highlights the position of OmV2. Circles at the nodes represent bootstrap support of 50 to 70% (white), 70 to 90% (gray), and 90 to 100% (black). The scale bar shows substitutions per site. (C) Cladogram representing the relationships between the fully sequenced prasinovirus clades from 85 core genes. Colored circles mark the branching positions of the percentage of the OmV2 orthologs.
