Fig. 3. cPRC1 and vPRC1 maintain H2AK119ub1 at “shared” Polycomb target genes.
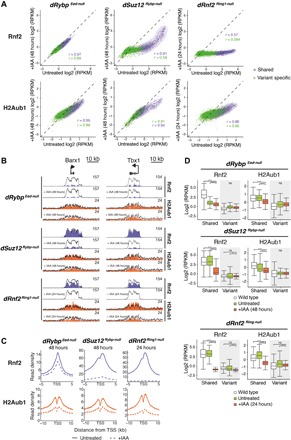
(A) Density scatterplots compare ChIP-seq signals of Rnf2 (top, ±5 kb around TSS) and H2AK119ub1 (H2Aub1) (bottom, ±10 kb around TSS) in untreated and IAA-treated dRybp Eed-null (48 hours, left), dSuz12 Rybp-null (48 hours, middle), and dRnf2 Ring1-null (24 hours, right) mESCs. r, Pearson’s correlation coefficient; “shared” TSS signal, violet; “variant-specific” TSS signal, green. (B) Genomic screenshots of Rnf2 (violet) and H2AK119ub1 (H2Aub1, orange) enrichments at two representative “shared” Polycomb target genes in untreated and IAA-treated dRybp Eed-null, dSuz12 Rybp-null, and dRnf2 Ring1-null mESCs. Superimposed are levels in wild-type mESCs (black line). (C) Meta plots of Rnf2 (top) and H2AK119ub1 (bottom) enrichments at “shared” Polycomb target genes in untreated and IAA-treated dRybp Eed-null, dSuz12 Rybp-null, and dRnf2 Ring1-null mESCs as indicated. (D) Box plots compare ChIP-seq signal of Rnf2 (left, ±5 kb around TSS) and H2AK119ub1 (H2Aub1) (right, ±10 kb around TSS) at “shared” and “variant-specific” Polycomb target genes in wild-type mESCs (white), untreated (green), and IAA-treated (red) dRybp Eed-null, dSuz12 Rybp-null, and dRnf2 Ring1-null mESCs. Significance (P value) was calculated using Wilcoxon rank test (****P < 2.2 × 10−16; ns, not significant).
