Fig. 6. Combined Pcgf1 and Suz12 depletion reduces H2AK119ub1, triggers activation of lineage-specific genes and results in loss of mESC self-renewal.
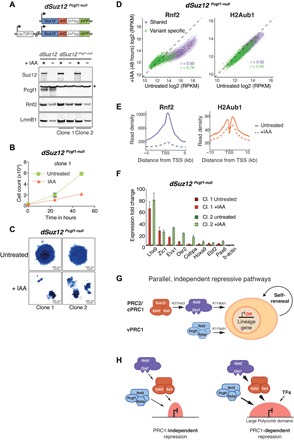
(A) Schematic representation of dSuz12 Pcgf1-null mESCs. Pcgf1-null mutation in mESCs expressing osTIR1 and endogenous Suz12 fused to AID-3×FLAG-GFP. Western blot of dSuz12 mESCs and two independent clones of dSuz12 Pcgf1-null mESCs show IAA-dependent fusion protein degradation and confirm Pcgf1 LOF mutation. * indicates unspecific band. Lamin B1 (Lmn B1) serves as loading control. (B) Proliferation assays of two independent clones of dSuz12 Pcgf1-null mESCs grown under serum conditions for 72 hours without (untreated) or with 250 μM IAA. Live cells were quantified by flow cytometry every 24 hours. Displayed is the median and SD of four replicate measurements starting at 24 hours IAA (time point, 0 hours). (C) Representative image of AP staining of dSuz12 Pcgf1-null mESC colonies cultured in the absence (untreated) or presence or 250 μM IAA for 5 days (+IAA). (D) Density scatterplots compare ChIP-seq signals of Rnf2 (left, ±5 kb around TSS) and H2AK119ub1 (H2Aub1) (right, ±10 kb around TSS) in dSuz12 Pcgf1-null mESCs without and with IAA treatment for 48 hours. r, Pearson’s correlation coefficient; “shared” TSS signal, violet; “variant-specific” TSS signal, green. (E) Meta plots of Rnf2 (left) and H2AK119ub1 (right) enrichments at “shared” Polycomb target genes in untreated and IAA-treated dSuz12 Pcgf1-null mESCs for 48 hours. (F) Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis compares expression of panel of “shared” Polycomb target genes in two independent clones of dSuz12 Pcgf1-null mESCs without and with IAA treatment for 48 hours. Gene expression is normalized to GAPDH and is shown relative to the average expression in untreated cells for each individual cell line. Error bars show SD (n = 2). (G) Cartoon illustrates model of parallel, independent mechanisms of PRC2/cPRC1 and vPRC1 targeting to ensure robust repression of lineage-specific genes and safeguard against exit from mESC pluripotency and spontaneous differentiation. (H) Cartoon displays shared PcG target genes embedded in small (left) or large (right) Polycomb repressive chromatin domains (red). PRC1 is critical for repression of PcG target genes located within large Polycomb domains marked by relatively high levels of PRC2/cPRC1 and vPRC1, as well as repressive histone modifications. Silencing of TSSs with smaller Polycomb domains is PRC1 independent and may be buffered by additional repressive chromatin modifications or the lack of activating transcription factors (TFs).
