Fig. 1.
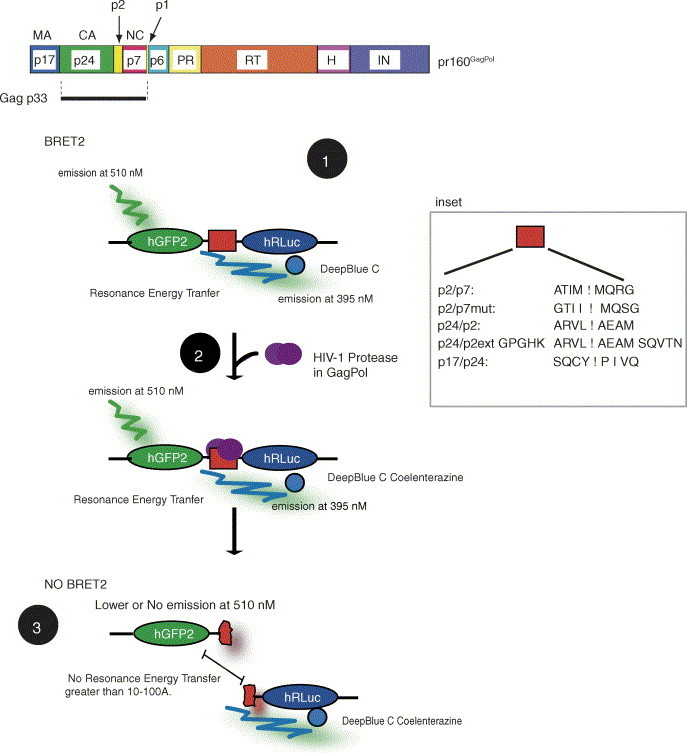
pr160GagPol is cleaved by the viral PR to generate mature porteins indicated in the scheme in A. The pr160GagPol-derived proteins are shown [MA (p17); CA (p24); spacer peptide, p2; NC (p7); spacer peptide, p1; PR, protease; RT, reverse transcriptase; H, RNAseH; IN, integrase). PR cleavage sites are located between each of the pr160GagPol mature proteins listed above. The p2/p7 site is located between p2 and p7 (NC). A scheme of the BRET2 assay is shown in B as described in the text. Briefly, the addition of DeepBlueC Coelenterazine initiates the resonance energy transfer to hGFP2 in the case of a close physical interaction or in the context of a hGFP2-hRLuc fusion protein (Step 1). HIV-1 PR will cleave its substrate (Step 2) to distance the donor and acceptor molecules thereby lowering the RET (Step 3). Inhibitors of PR such as Vif (this manuscript), or PR inhibitors used in HAART will maintain the BRET2 signal because PR activity is inhibited. The inset shows the PR cleavage sites and mutant sites introduced between the donor and acceptor molecules tested in this report. The letter codes for amino acids are indicated.
