Fig. 1. Dynamics of bead displacement and chromatin deformation with increasing force frequency.
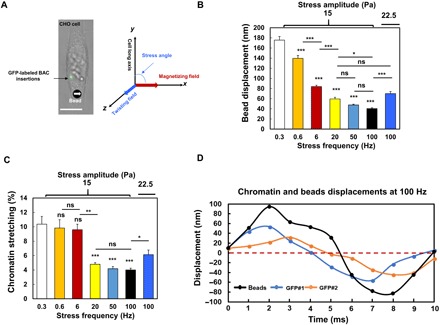
(A) Left: Bright-field image of a DHFR-deficient Chinese hamster ovary (CHO) cell labeled with LacI-GFP spots in the same chromatin domain stably transfected with BAC Lac operator (LacO) mouse DHFR transgenes. An RGD (Arg-Gly-Asp)–coated magnetized ferromagnetic bead (bottom, the black dot with a white arrow) was bound to the cell surface via integrins. Right: The magnetic bead was magnetized along the x axis with a 1000-Pa pulse (<1 ms), and a sinusoidal twisting field was applied along the z axis to rotate the bead to apply rotational stresses to the cell. The cell’s long axis was along the y axis or at an angle (0° to 90°) (defined as stress angle) to the x axis. Changes in green fluorescent protein (GFP) spots distances were used for quantification of chromatin deformation. Scale bar, 10 μm. (B) Displacements of center of magnetic beads on the cell surface by a 15-Pa sinusoidal stress at various frequencies; n = 60 cells for each frequency; n = 31 cells for 22.5-Pa bead stress at 100 Hz. (C) Chromatin stretching in response to 15-Pa stress at different frequencies. n = 55, 27, 31, 31, 46, and 43 cells for 0.3, 0.6, 6, 20, 50, and 100 Hz, respectively. n = 78 cells for 22.5-Pa stress at 100 Hz. (D) Representative original data of displacements of beads and GFP spots with stress application at 100 Hz. The nonperfect sinusoidal waves of the GFP curves are likely due to the baseline noises of GFP spot spontaneous movements that are 2.3 ± 2.0 nm. Mean ± SEM for subfigures (B and C); n ≥ 3 independent experiments; ns, not statistically significant; *P < 0.05, **P < 0.01, ***P < 0.001; one-way analysis of variance (ANOVA) test with Bonferroni modifications.
