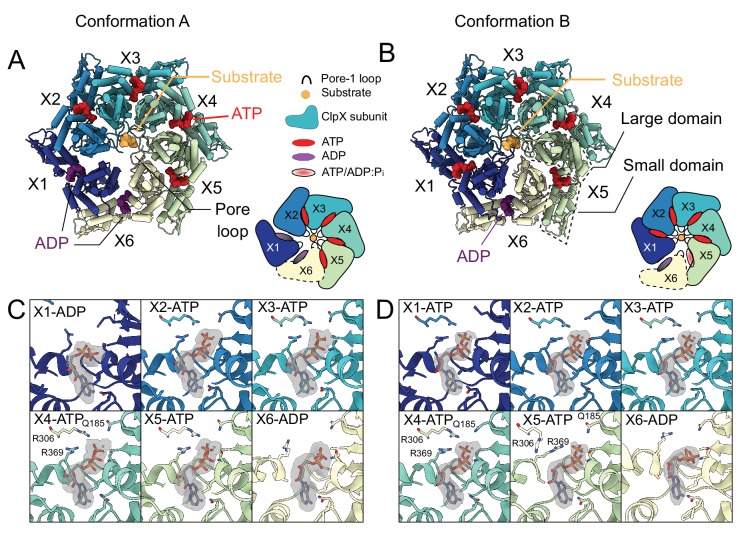
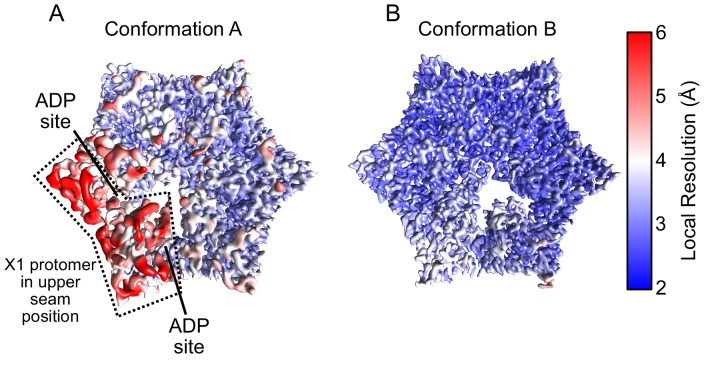
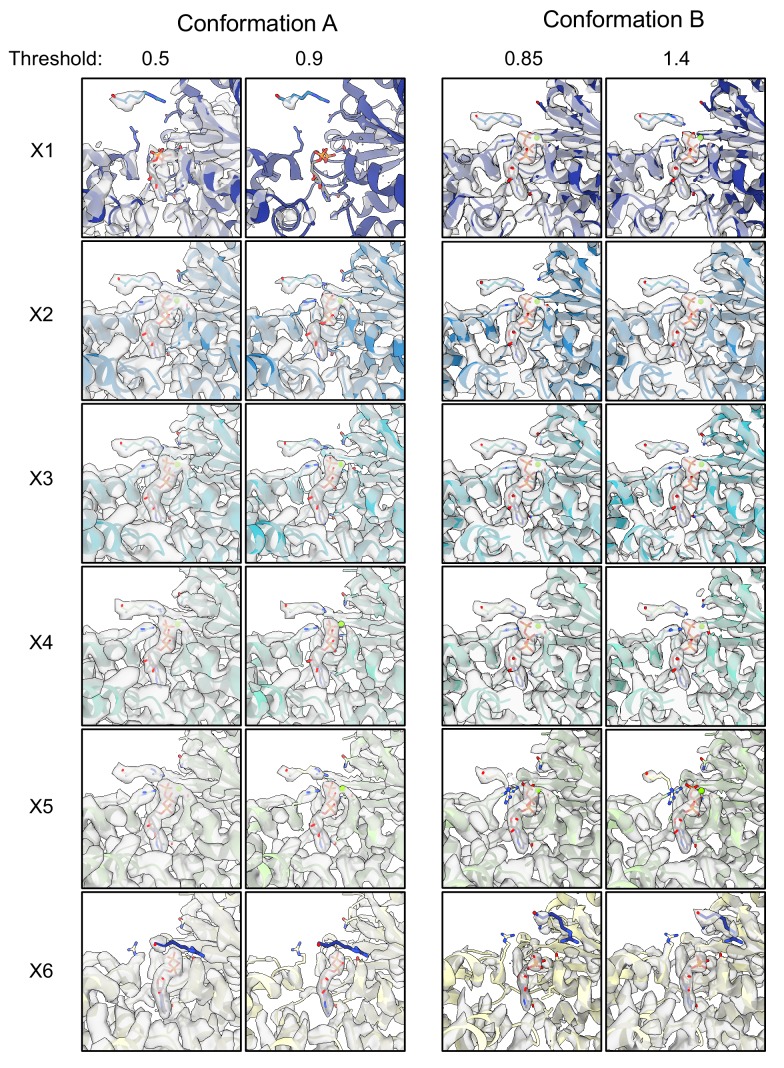
Figure 4. Nucleotide occupancy and interactions with ClpX.
(A and B) Model of ClpX in conformations A and B, looking into the ClpX pore. Nucleotides are shown and color-coded, with ATP red and ADP purple, and bind between the large and small domains (boxed). Inset shows a schematic of the protomer positions and nucleotide occupancies. Conformation A has two bound ADP, in the X1 (dark blue, US in Conformation A in this representation) and X6 (yellow, LS in Conformation B in this representation) protomers, with the X1 protomer disengaged from the substrate and away from the axial pore (US position). In Conformation B only a single ADP is bound, protomer X1 is reengaged with substrate and X6 has disengaged. (C) Nucleotide-binding sites of Conformation A, with key interacting residues shown along with the experimental density maps corresponding to bound nucleotide. In the ADP-bound sites, both the arginine finger (R306) and the sensor-II arginine (R369) have moved away, while in the ATP bound sites they form close contacts with the β and γ-phosphates. (D) Nucleotide binding in Conformation B. As in Conformation A, R306 and R369 have moved away from the ADP, while in the X5 protomer only the arginine finger has moved away, while the sensor-II transitions closer to the γ-phosphate.