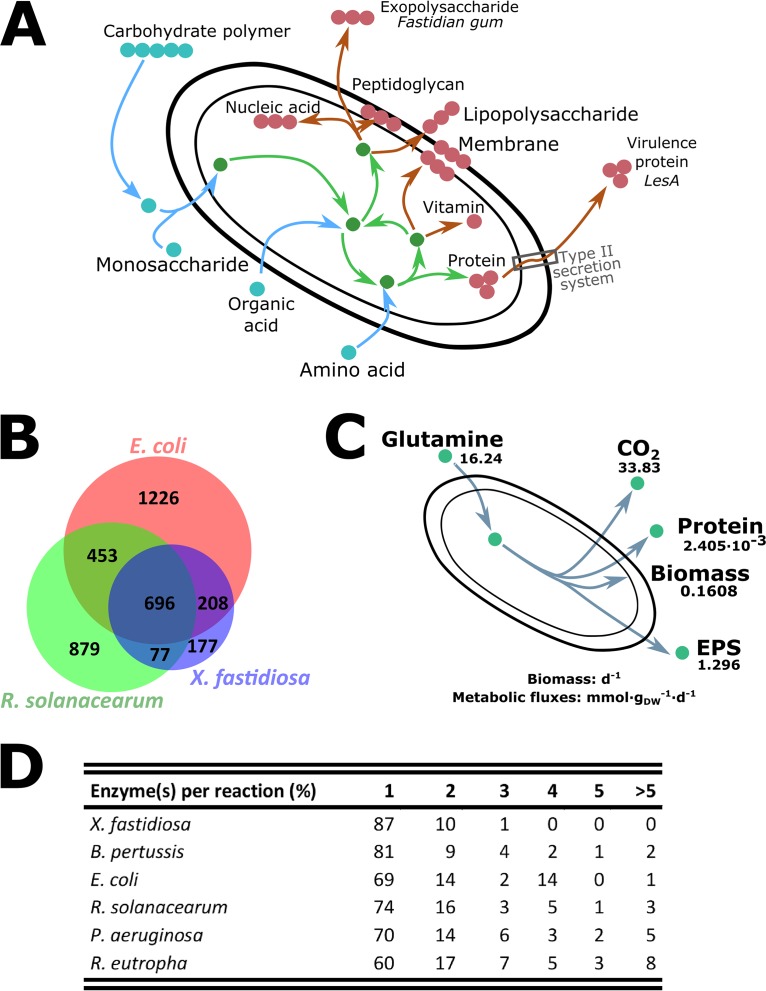
FIG 1.
General characteristics of the reconstructed metabolic network. (A) Global overview of the reconstructed metabolic pathways in Xylella fastidiosa. This schema highlights the metabolic behaviors of Xylella fastidiosa. For catabolism (external substrates and catabolic reactions in blue), X. fastidiosa is able to degrade polymers and assimilate them or catabolize elements from xylem sap such as amino acids and organic acids. Central metabolites and reactions are schematized in green (glycolysis and TCA cycle). Metabolites/reactions in red/brown highlight anabolic capabilities: the production of biomass macromolecules (membranes, lipopolysaccharides, peptidoglycans, vitamins, proteins, and nucleic acids) and the secretion of virulence factors (virulence protein through the type II secretion system and fastidian gum [EPS]). (B) Venn diagram depicting the number of metabolic reactions in E. coli, R. solanacearum, and X. fastidiosa. The lists of the reactions found in each part of the Venn diagram are available in Data Set S2. (C) Inputs and outputs of an FBA simulation of X. fastidiosa growth on glutamine under conditions close to experimental conditions. The objective function of FBA was minimization of glutamine import. Growth and production rates were calculated from previous publications (19, 82). Flux values are in mmol·g (DW)−1·day−1 for all inputs/outputs except for biomass (day−1). See Text S3 for the constraints used for the simulation and Data Set S2 for the FBA solution. Scripts are available on https://github.com/lgerlin/xfas-metabolic-model. (D) Distribution of the number of enzymes for each metabolic reaction in X. fastidiosa and metabolic models from other bacteria. Proportions of reactions carried out by 1, 2, 3, 4, 5, and >5 enzymes were computed for each metabolic model. The complete data are available in Data Set S2.