Figure 1: Batf is expressed by lung ILC2 cells and is predicted to regulate distinct gene sets in KLRG1pos populations.
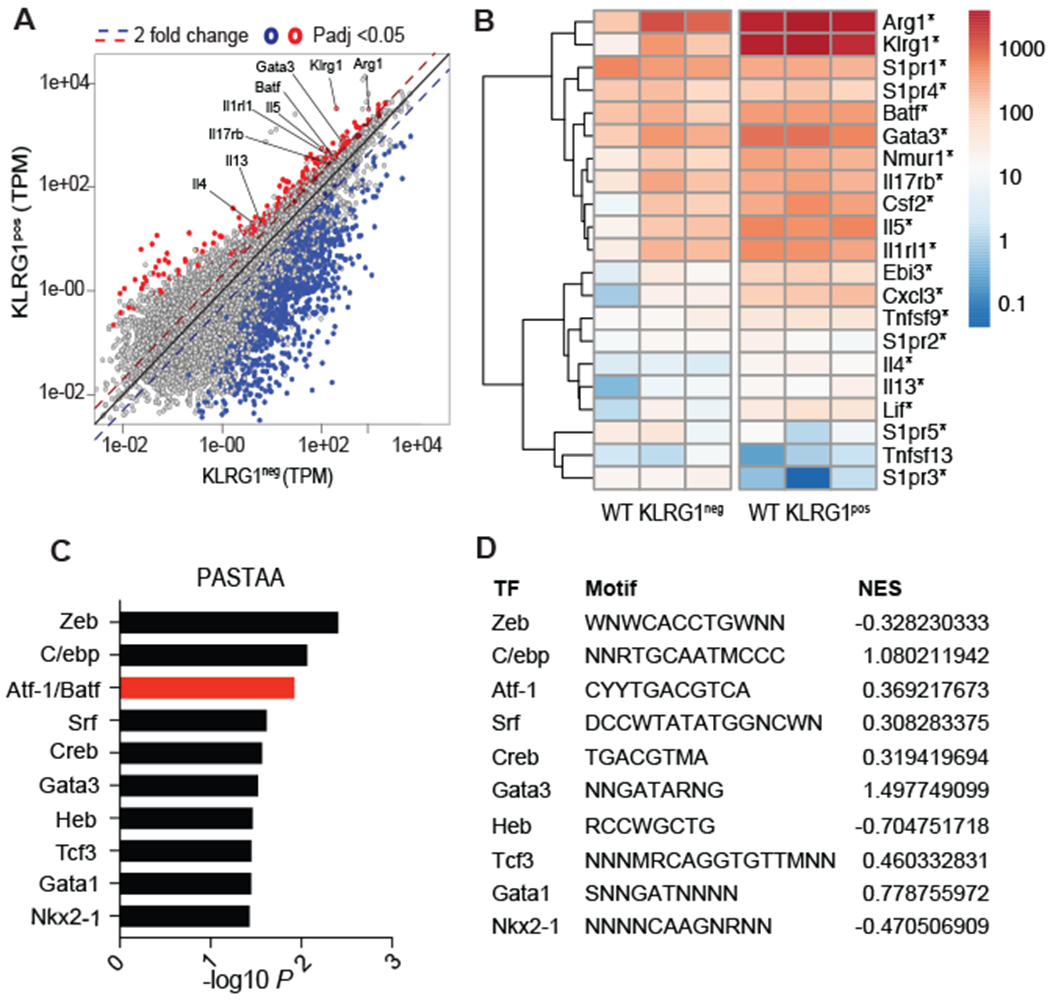
WT C57BL/6 mice were injected intraperitoneally (i.p.) with IL-33 daily for 4 days and pulmonary KLRG1neg (non-ILC2s) and KLRG1pos (ILC2s) were sorted on day 5 and used for RNA-sequencing. (A) Diagonal scatter plot comparing differentially expressed genes (padj <0.05) among KLRG1neg and KLRG1pos cells. (B) Heat map of ILC2-related genes as well as S1P receptor genes upregulated in KLRG1pos ILCs *p≤0.05. (C) Transcription factors predicted by PASTAA analysis to be involved in regulating genes that were enriched among KLRG1pos, compared to KLRG1neg, populations. (D) Normalized enrichment scores of the predicted transcription factors as assessed with gene set enrichment analysis of KLRG1pos ILC2s compared to the KLRG1neg population. Binding motifs as reported by TRANSFAC are also included in the table where S=C or G, W=A or T, R=A or G, Y=C or T, M= A or C, and N=any base. RNA-sequencing analysis was compiled from three separate experiments with n=3-6 mice per group.
