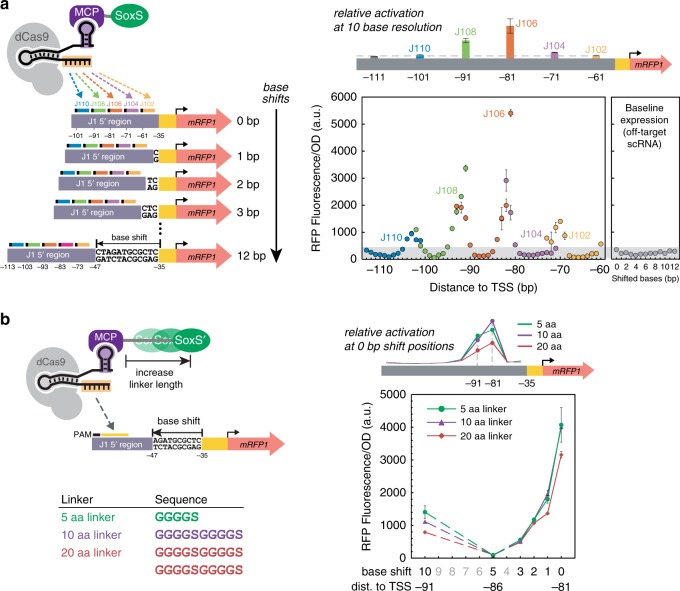
Fig. 4. CRISPRa is sensitive to the precise position of the scRNA target.
a CRISPRa displays periodic positioning dependence with peak activities every 10–11 bases between −60 and −100 from the TSS. Reporter genes were constructed by inserting 0–12 bases upstream of the −35 region of the J1-J23117-mRFP1 reporter. Five scRNA sites (J102, J104, J106, J108, J110) with positions −61, −71, −81, −91, −101 from the TSS on the non-template strand of the original promoter were targeted. In this way, the complete −61 to −113 region can be covered at single base resolution. The color coding indicates data for the same target site shifted across a 12 base window. The panel on the right shows the baseline expression of reporters with shifted bases when an off-target scRNA was used (J206). The gray area represents the range of the baselines among the reporter series. For comparison, previous CRISPRa data for the J102, J104, J106, J108, J110 target positions at 10 base resolution are shown on the schematic above the plot3. b Extending the linker length between MCP and SoxS does not change the position dependence of CRISPRa. The J1-J23117-mRFP1 reporter plasmid series with base shifts were delivered together with CRISPRa components for targeting J106. The MCP-SoxS(R93A) effector contained 5aa, 10aa, and 20aa linkers. For comparison, previous CRISPRa data with 5aa, 10aa, or 20aa linker between MCP and SoxS targeting at the −81 and −91 positions are shown on the schematic above the plot3. Values in a and b represent the average±standard deviation calculated from n = 3 biologically independent samples. Source data are provided as a Source Data file.