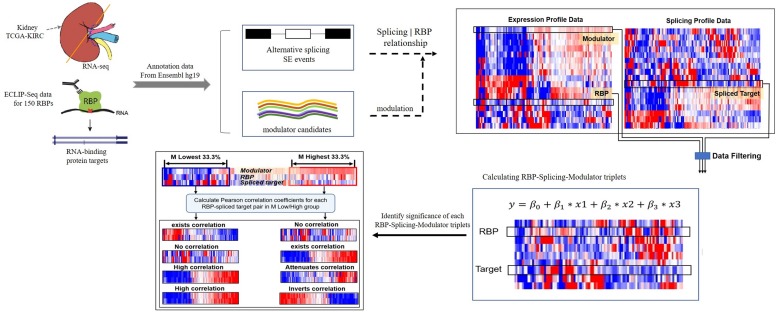
FIGURE 1.
A schematic diagram of workflow. Briefly, the dataset in this study derived from TCGA-KIRC and ENCODE ECLIP-Seq, hg19 was used as reference gene annotation from Ensemble. Each triplet contains three object: RBP, target and a modulator. Gene expression level is the input of RBP and modulator candidates, splicing outcomes (PSI value) is used to estimate the splicing level of target. Data filtering criteria as follows: (1) log2 (CPM) ≥ 1 (2) remove events with “NA” samples > 100 (3) CV(PSI) > 0.1. Then using the linear regression model to predict triplets. Only the triplets with significant β3 p-value will be considered and selected to the following analysis. Finally, for each triplet, we group the samples into “low” and “high” groups based on the expression level of modulator (bottom/top 33% samples) in the specific triplet, and we compare the Pearson correlation coefficient values of RBP expression and target PSI value in two groups, identify the modulator function categories.