Figure 4.
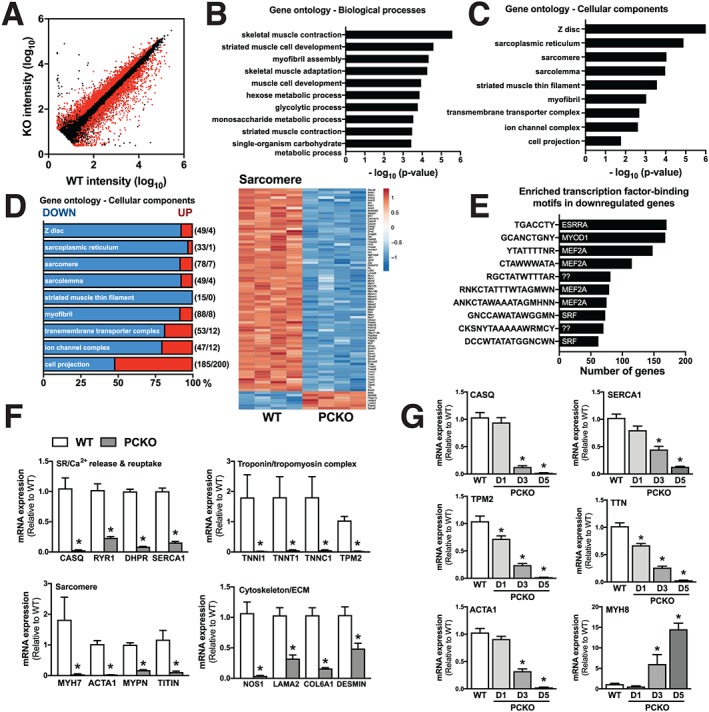
Down‐regulation of major muscle gene programs in PCKO mice. (A) Scatter plot of gene expression profiles in WT and PCKO muscles. The x and y axes indicate the gene expression levels (log10 transformation). Red dots represent differentially expressed genes in PCKO vs. WT muscle [n = 4 WT, n = 4 PCKO; false discovery rate (FDR) < 0.1]. (B–C) Top 10 overrepresented gene ontology (GO) terms (local FDR < 0.1) for differentially expressed genes in PCKO vs. WT muscle, (B) biological processes and (C) cellular components. (D) Expression profile [down (blue) and up (red)] for genes associated with cellular components GO categories in panel C. Numbers in parentheses indicate the number of altered genes (DOWN/UP, FDR < 0.1). Heat map generated using the ClustVis online tool using log10 transformed probe set intensities for genes differentially expressed (FDR < 0.1) in PCKO vs. WT mice and associated with the GO:cellular components categories ‘GO:0030017, sarcomere’. The colour ranges from deep red (high abundance) to deep blue (low abundance); white is no change. (E) Top 10 transcription factor binding motifs enriched in genes down‐regulated (FDR < 0.1) in PCKO vs. WT mice. Bar graph lists enriched motifs (left) and the transcription factors (TFs) predicted to be associated with these motifs (right). Question marks indicate that the motif is not associated with a TF. (F) mRNA levels of indicated genes in quadriceps normalized to Tbp, in D5 PCKO mice vs. WT mice (n = 6 WT and n = 6 PCKO). (G) mRNA levels of indicated genes in gastrocnemius muscle normalized to Tbp, in D1, D3, and D5 PCKO mice compared with WT mice (n = 12 WT, n = 6 D1 PCKO, n = 6 D3 PCKO, and n = 6 D5 PCKO). Statistics: Data reported as means ± SEM. (F) Student's t‐test, * P < 0.05, PCKO vs. WT. (G) One‐way analysis of variance, Bonferroni post hoc test, * P < 0.05, PCKO vs. WT.
