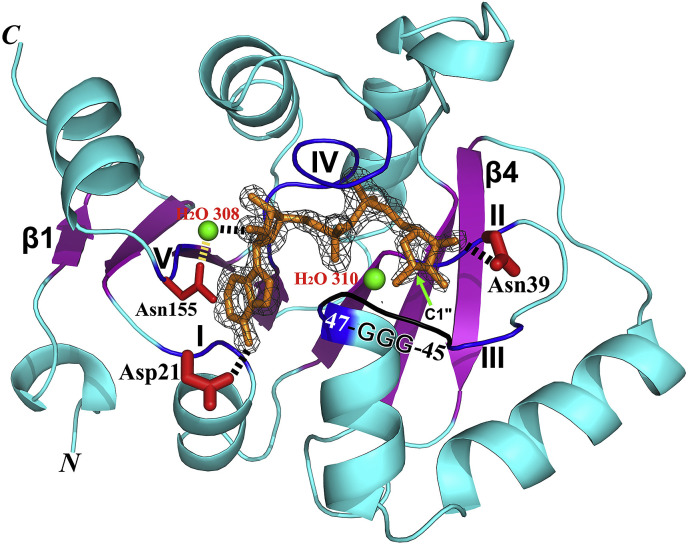
Fig. 4.
Structure of the MERS-CoV macrodomain I (Mac1, X domain) in complex with ADP-ribose (ADPr) (PDB entry: 5HOL). The protein features an α/β/α sandwich fold. The central β sheet with the strand order β1−β2−β7−β6−β3−β5−β4 is shown in purple, β1 and β4 are labeled. An Fo-Fc omit difference map of ADPr is shown in black (contoured at 4.0 σ). The ADPr itself is displayed as brown sticks. The five regions (blue) relating to ADPr binding are marked by Roman numbers I – V. Fixing the two ends of the ADPr, Asp21 and Asn39 are displayed by thicker red sticks. The O2′ of ADPr forms a hydrogen bond with a water molecule (H2O 308; green sphere) being stabilized by the side-chain of Asn155. The “GGG” triple-glycine motif is displayed in black. H2O 310 (green sphere) corresponds to a water molecule that has been proposed to mediate a nucleophilic attack onto the C1″ atom of the ADPr in the de-MARylation reaction catalyzed by the VEEV X domain (Li et al., 2016a). The N and C termini of the X domain are marked. This figure and Fig. 6 were prepared using Pymol (Schrödinger; http://www.pymol.org/).