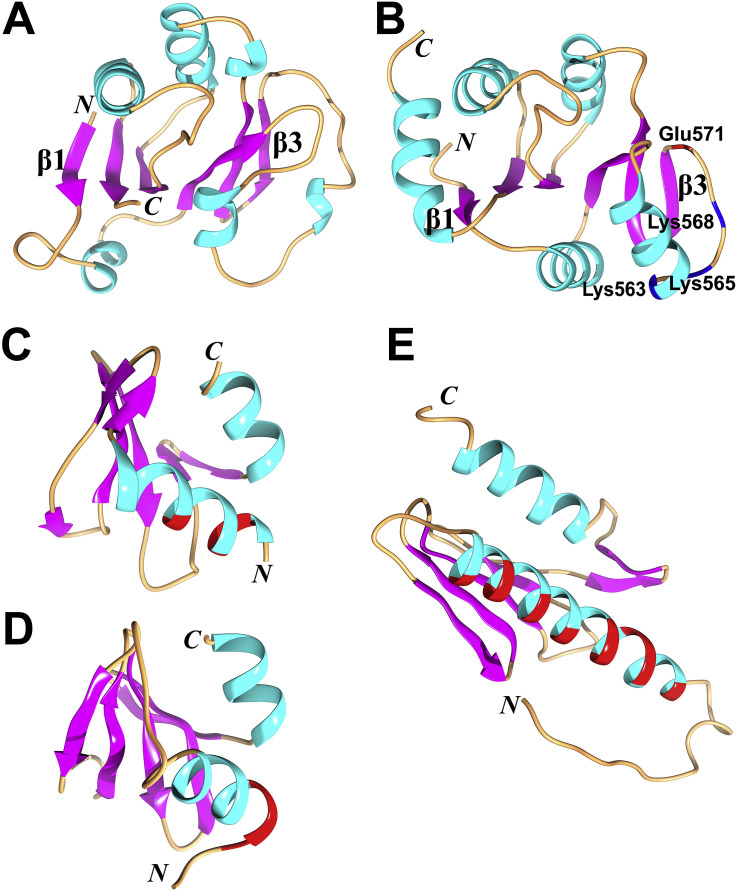
Fig. 5.
Structures (in cartoon style) of the macrodomains II (Mac2) and III (Mac3), of the Domain Preceding Ubl2 and PL2pro (DPUP) of SARS-CoV and MHV, as well as of the frataxin-like fold protein Yfh1. (A) and (B) Mac2 and Mac3 (PDB entry: 2W2G; Tan et al., 2009). Both domains possess the α/β/α sandwich fold. The central six β strands in the order β1−β6−β5−β2−β4−β3 are displayed in purple. A predominantly positively charged surface patch (Lys563+Lys565+Lys568+Glu571; Nsp3 numbering) of Mac3 being involved in binding oligo(G) (Kusov et al., 2015) is labeled. (C) The SARS-CoV DPUP NMR structure (PDB entry: 2KQW; Johnson et al., 2010). (D) The MHV DPUP X-ray crystal structure (PDB entry: 4YPT; Chen et al., 2015). (E) Structure of the yeast frataxin-like protein Yfh1, as determined by NMR spectroscopy (PDB entry: 2GA5; He et al., 2004). All structures shown in (C), (D), and (E) display the typical frataxin-like fold. Two α helices located at the N- and C- terminal of each structure form one plane and the β sheet forms the other plane. The negatively charged residues (Asp or Glu) in the first α helix (α1) are shown in red (in (C), (D), and (E)); they are possibly involved in binding metal ions. The N and C termini of all structures are marked.