Fig. 3.
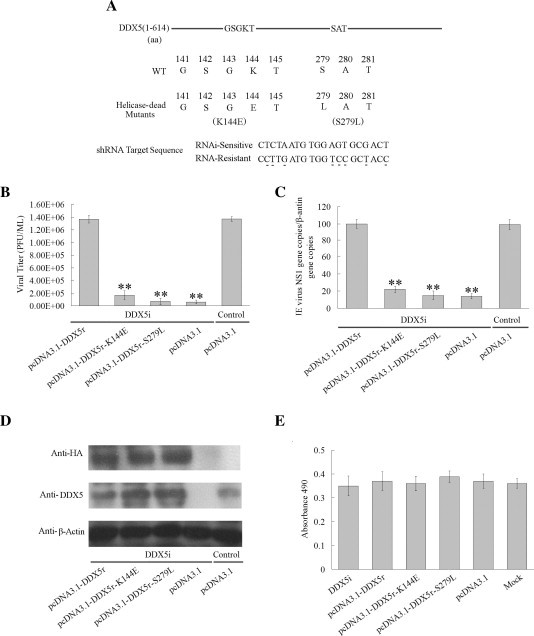
The effect of exogenous DDX5 and helicase-dead DDX5 on JEV replication. (A). The residues of DDX5 helicase-dead mutants (K144E, S279L) were shown. The sequences of DDX5 specific-shRNA and the shRNA-resistant targeting region were aligned. The different nucleotides are underlined. (B). BHK-21 cells were first transfected with DDX5i plasmid. 24 h later, the cells were transfected the pcDNA3.1-DDX5-HA, pcDNA3.1-DDX5r-K144E-HA, pcDNA3.1-DDX5r-S279L -HA or pcDNA3.1 plasmid, viral titers determined by plaque assay at 48hpi (MOI = 0.01). (C). The level of JEV genome RNA was detected by real-time PCR in the cells infected with JEV after first transfected with the DDX5i plasmid then with pcDNA3.1-DDX5r-HA, pcDNA3.1-DDX5r-K144E-HA or pcDNA3.1-DDX5r S279L-HA, the untargeted shRNA and pcDNA3.1 plasmid were used as the control. (D). The expression of transfected DDX5 mutants plasmids and the endogenous DDX5 were detected by Western blot using anti-HA monoclonal antibody and anti-DDX5 polyclonal antibody. The β-actin was used as loading control. (E). The cell viability of BHK-21 cells transfected with DDX5i, HA-DDX5 or DDX5 mutant plasmids was detected by CytoTox 96® NonRadioactive Cytotoxicity Assay (Promega) at 72 h after transfection. The differences between means were considered significant at ∗p < 0.05, very significant ∗∗p < 0.01.
