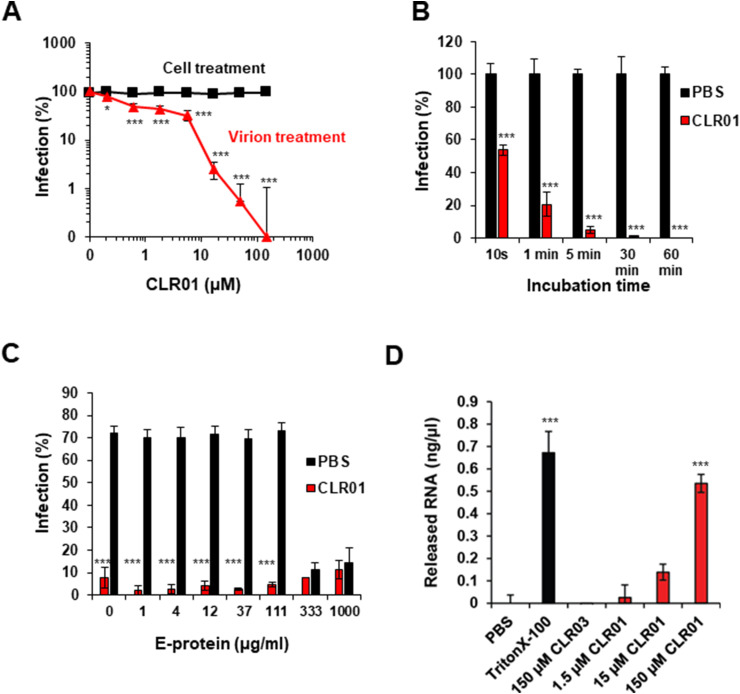
Fig. 3.
Mechanism of CLR01 inhibition of ZIKV infectivity. (A) For virus treatment, ZIKV was incubated with CLR01 for 10 min at room temperature before the mixtures were added to Vero E6 cells. For cell treatment, CLR01 was added directly to the cells; after 2 h, the medium was replaced and the cells were infected with ZIKV MR766. 2 dpi, cell-based ZIKV immunodetection assay was performed. Values represent mean ± SD (n = 3). One-way ANOVA (non-parametric, grouped), followed by Bonferroni's multiple comparison tests were applied to compare the different CLR01/CLR03 concentrations to cells infected in the absence of compound (* denotes p < 0.01; *** denotes p < 0.0001) (B) ZIKV was incubated for the indicated times with PBS or 10 μM CLR01 before the mixture was added to Vero E6 cells. 2 dpi, cell-based ZIKV immunodetection assay was performed. Values represent mean ± SD (n = 3). One-way ANOVA (non-parametric, grouped), followed by Bonferroni's multiple comparison tests were applied to compare cells infected with CLR01-treated ZIKV to cells infected with ZIKV that had been incubated with PBS for the same time period (*** denotes p < 0.0001). (C) Indicated concentrations of ZIKV E protein were titrated to 100 μM CLR01 or PBS before ZIKV was added. Mixtures were used to inoculate Vero E6 cells. 3 dpi, the number of adherent, viable cells were determined using the MTT assay (Müller et al., 2016). The values shown are mean ± SD from triplicate infections. One-way ANOVA (non-parametric, grouped), followed by Bonferroni's multiple comparison tests were applied to compare cells treated with different concentrations of ZIKV E protein and CLR01 to cells that had been treated with the same concentrations of E protein and PBS (*** denotes p < 0.0001). (D) ZIKV MR766 was incubated with PBS, 150 μM Triton X-100, 1.5-150 μM CLR01 or 150 μM CLR03 for 30 min at 37 °C. Samples were inactivated by UV light of a laminar flow for 60 min. Then, 10 μl of the samples were used to determine RNA concentrations using the QuantiFluor® RNA System and a Quantus Fluorometer (Promega). Background values obtained from control samples using cell supernatant of uninfected cells were subtracted from the respective signals. RNA levels of virus stock incubated with PBS were subtracted from these values. Data points represent mean ± SD (n = 3). One-way ANOVA (non-parametric, grouped), followed by Bonferroni's multiple comparison tests were applied to compare samples treated with different CLR01 concentrations, CLR03 or Triton X-100 to PBS-treated virus (*** denotes p < 0.0001).