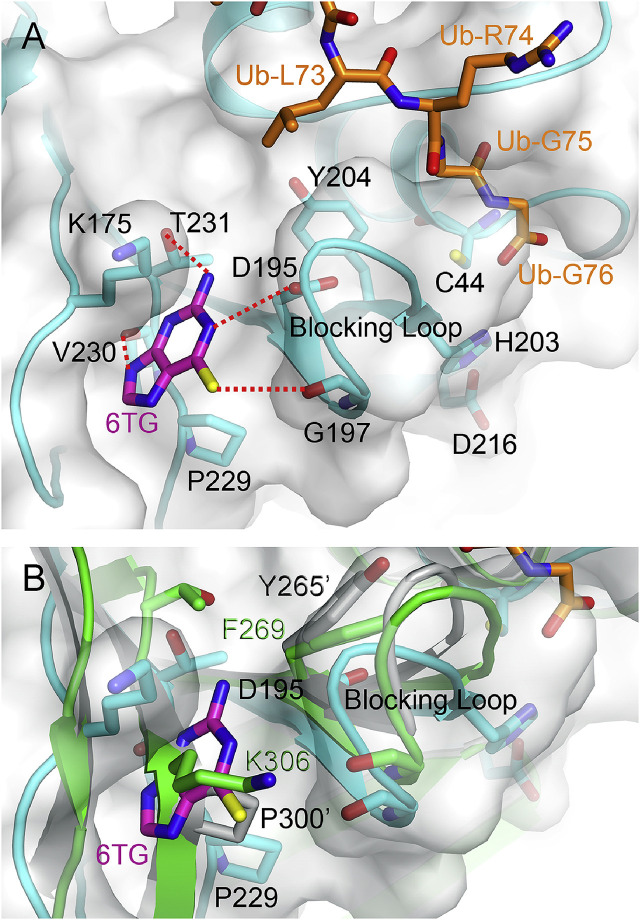
Fig. 4.
Model structure of PEDV PL2proin complex with 6TG. (A) The model structure of PEDV PL2pro (cyan) was generated by SWISS-MODEL (Arnold et al., 2006). The location of Ub (orange) is based on overlaying with the complex structure of MERS-CoV PLpro C111S – Ub (PDB code: 4WUR) (Lei and Hilgenfeld, 2016). The docking of 6TG (magenta) was performed using AutoDock Vina. The residues are shown as sticks while the putative polar interactions are shown as dotted lines. (B) Overlay of the docked model, SARS- (grey; PDB code: 4M0W) and MERS-CoV (green) PLpro-Ub complex structures. The same location is occupied by the residue Pro300 of SARS-CoV PLpro and the residue Lys306 of MERS-CoV PLpro, respectively, suggesting that 6TG cannot bind to this site in the two enzymes. The figures were produced using PyMol (http://www.pymol.org). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)