Fig. 2.
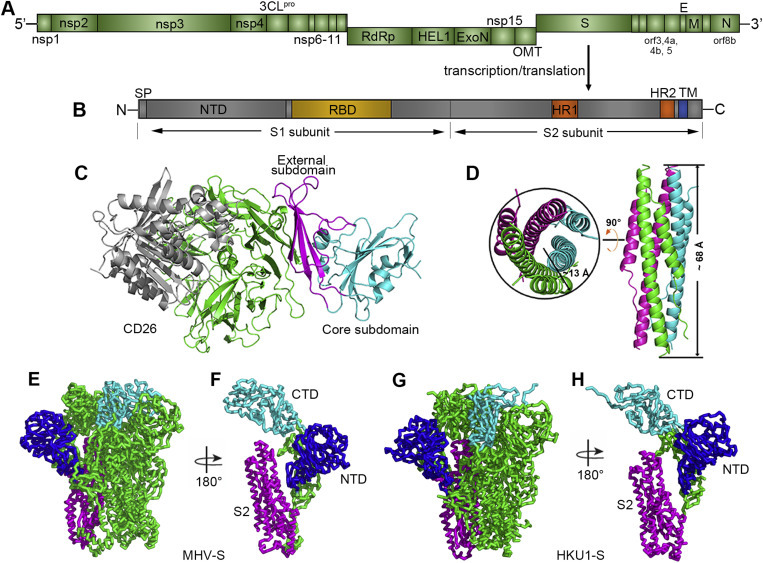
Genome arrangement of MERS-CoV and structure of the spike protein-receptor complex. (A) Schematic representation of the MERS-CoV genome. Abbreviations: nsp, nonstructural protein; 3CLpro, 3C-like protease; RdRp, RNA-dependent RNA polymerase; HEL1, superfamily 1 helicase; ExoN, 5′-3′ exonuclease; OMT, S-adenosylmethionine-dependent ribose 2′-O-methyltransferase; S, spike protein; orf, open reading frame; E, envelope protein; M, membrane protein; and N, nucleocapsid. In addition, PLpro (papain-like protease) is located in nsp3, and NendoU (nidoviral endoribonuclease specific for U) is located in nsp15. The gene coding for accessory protein orf8b overlaps with the N-coding gene (Cotten et al., 2013). (B) Schematic representation of the MERS-CoV spike protein. Abbreviations: SP, signal peptide; NTD, N-terminal domain; RBD, receptor binding domain; HR1/2, heptad repeat 1/2; and TM, transmembrane domain. (C) Complex structure between the MERS-RBD and its receptor CD26. The core and external subdomains are highlighted in cyan and magenta, respectively, while the receptor is colored in green for the β-propeller domain and in gray for the α/β-hydrolase domain, respectively. (D) Crystal structure of the HR1/HR2 fusion core. The three HR1/HR2 chains are colored in green, cyan, and magenta, respectively. The approximate size of the bundle is indicated. The left panel represents the top view, and the right panel represents the side view. The figure was used upon approval of the authors in Gao et al., 2013. (E) and (G) Ribbon diagrams showing the overall structures of MHV-S and HKU1-S trimers, respectively. (F) and (H) Ribbon diagrams showing one MHV-S and HKU1-S molecule, respectively. NTD, CTD and S2 are colored in blue, cyan, and magenta, respectively in E-H.
