Fig. 1.
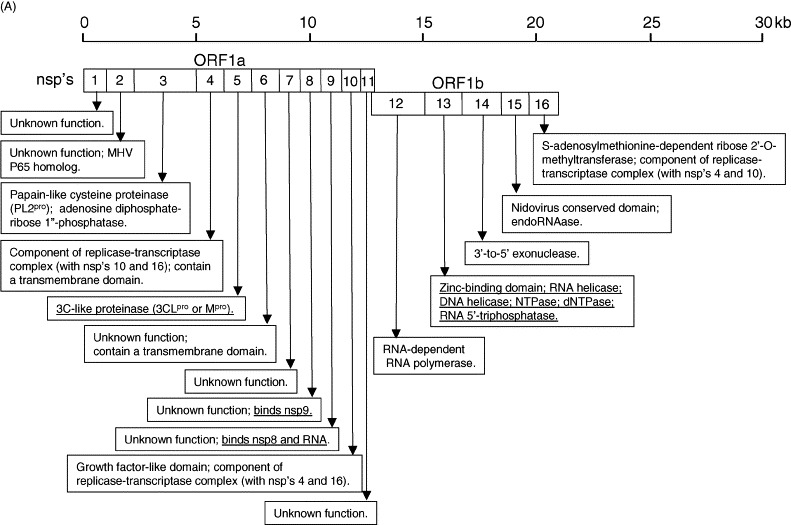
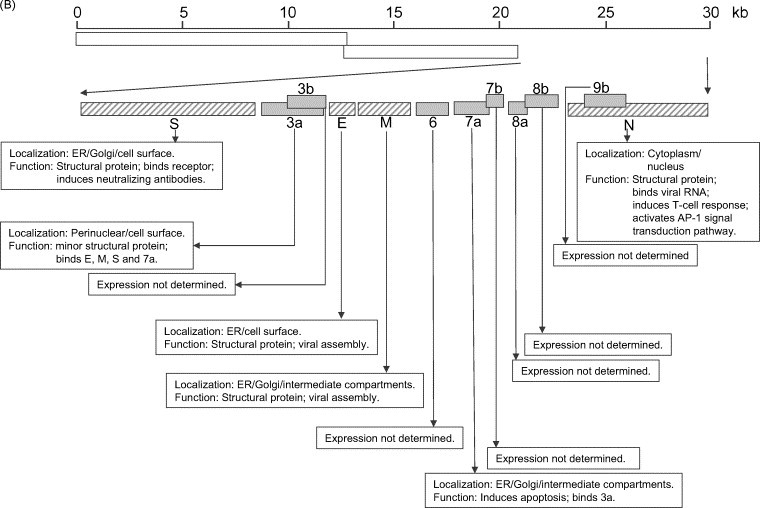
Summary of the SARS-CoV genome organization and viral protein expression. (A) Replicase genes (ORFs 1a and 1b), constituting the first 2/3 of the genome, translate into two large polyproteins, pp1a (486 kDa) and pp1ab (790 kDa). Expression of the ORF 1b-encoded region of pp1ab involves ribosomal frameshifting into the −1 frame just upstream of the ORF 1a translation termination codon. Proteolytic processings of these polyproteins are mediated by viral cysteine proteinases (nsp3, also called PL2pro and nsp5, also called 3CLpro or Mpro) resulting in 16 non-structural proteins (nsp's; open boxes). Putative functional domains present in each nsp's are shown in the text boxes. Functions that have been demonstrated with recombinant proteins are underlined. (B) Open reading frames (ORFs) in the remaining 1/3 of the genome are translated from eight subgenomic mRNAs. Four of these encode the structural proteins (checked boxes), spike (S), membrane (M) and envelope (E) and nucleocapsid (N). Another eight SARS-CoV-unique ORFs (grey solid boxes) encode putative “accessory” proteins with no significance sequence homology to viral proteins of other coronaviruses (3a, 3b, 6, 7a, 7b, 8a, 8b and 9b). The cellular localization and functions of some of these viral proteins have been demonstrated (see text boxes). Also note that S, E, M and ORF 6 are expressed from individual subgenomic mRNAs, while 3a and 3b are predicted to be produced from the same subgenomic mRNA. Similarly, 7a and 7b are also produced from one subgenomic mRNA, and 9b is produced from the same subgenomic mRNA as N. The expression of 3b, 7b and 9b may be via “leaky scanning” by ribosomes or involve a mechanism like internal ribosomal entry. However, it cannot be rule out that they may also be expressed from the synthesis of yet undetected additional subgenomic mRNAs.
