Fig. 4.
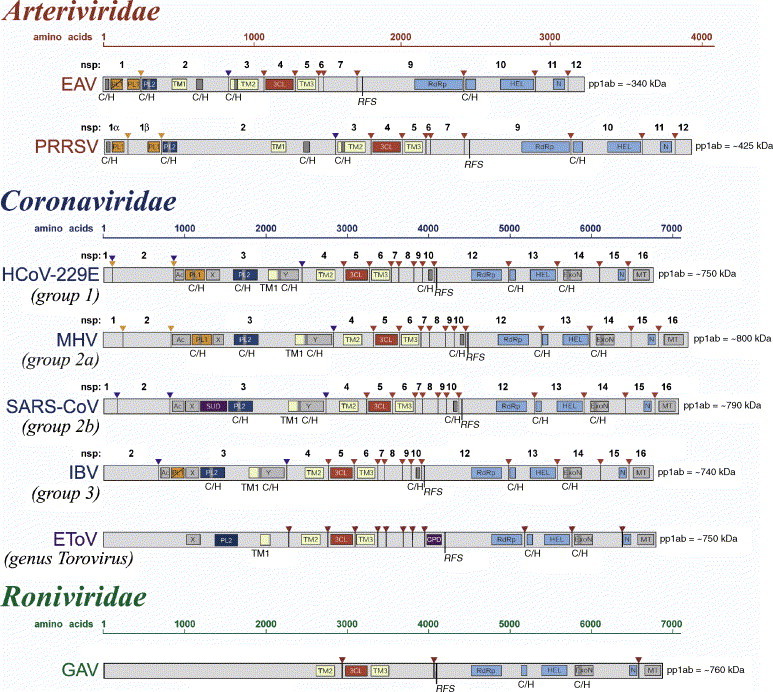
Domain organization of the replicase pp1ab polyprotein for selected nidoviruses. Shown are currently mapped domains in the pp1ab replicase polyproteins of viruses representing major lineages of nidoviruses. Arrows represent sites in pp1ab that are cleaved by papain-like proteinases (orange and blue) or chymotrypsin-like (3CLpro) proteinase (red). For viruses with the full cleavage site map available (arteriviruses and coronaviruses), the proteolytic cleavage products are numbered. A tentative cleavage site map for EToV is from an unpublished work by A.E. Gorbalenya. Within the cleavage products, the location and names of domains that have been identified as structurally and functionally related are highlighted. These include diverse domains with conserved Cys and His residues (C/H), putative transmembrane domains (TM), domains with conserved features (AC, X and Y), and domains that have been associated with proteolysis (PL1, PL2, and 3CL), RNA-dependent RNA synthesis (RdRp), helicase (HEL), exonuclease (ExoN), uridylate-specific endoribonuclease (N; NendoU in the main text), methyl transferase (MT) and cyclic phosphodiesterase (CPD) activities. Note that due to space limitations, the domain names in this figure may be abbreviated derivatives from those used in the main text. Polyproteins of large and small nidoviruses are drawn to different scales. This figure was updated from Fig. 39.4 presented in Siddell et al. (2005).
